













| General Information: |
|
| Name(s) found: |
par-1-PB /
FBpp0085647
[FlyBase]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 905 amino acids |
Gene Ontology: |
|
| Cellular Component: |
germline ring canal
[IDA]
cell cortex [IDA] fusome [IDA] basolateral plasma membrane [IDA] |
| Biological Process: |
regulation of cell shape
[IMP]
germarium-derived oocyte fate determination [IMP][TAS] axis specification [IMP] border follicle cell delamination [IMP] actin filament organization [IMP] gamete generation [IMP] border follicle cell migration [IMP] oocyte differentiation [TAS] microtubule cytoskeleton organization [IMP] protein amino acid phosphorylation [ISS][NAS] oocyte anterior/posterior axis specification [TAS] anterior/posterior axis specification [TAS] maintenance of protein location [TAS] regulation of pole plasm oskar mRNA localization [IMP] pole plasm protein localization [IMP] oocyte nucleus localization involved in oocyte dorsal/ventral axis specification [IMP] ovarian follicle cell development [IMP] pole plasm oskar mRNA localization [TAS] regulation of Wnt receptor signaling pathway [TAS] antimicrobial humoral response [IMP] oocyte microtubule cytoskeleton organization [IMP][TAS] |
| Molecular Function: |
tau-protein kinase activity
[IDA]
ATP binding [IEA] protein kinase activity [IDA] protein serine/threonine kinase activity [ISS][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTAMRTTLQ SVPEALPADS VSNGTASNVA APAAPVSSAT NAVPPLAAVS STTATYATNS 60
61 ISTSSHSVKD QQQQQQQQQH DSANANIVSL PPTTTPVANT NTMMPIVTSS NSATSNSTAA 120
121 TPTPASGAAA TGGVGSVSQG PATVSASAAN TNHSHQHSHQ HHHHVANNMT TDGARLSSNN 180
181 SAVVASSAIN HHHHHTPGSG VAPTVNKNVL STHSAHPSAI KQRTSSAKGS PNMQMRSSAP 240
241 MRWRATEEHI GKYKLIKTIG KGNFAKVKLA KHLPTGKEVA IKIIDKTQLN PGSLQKLFRE 300
301 VRIMKMLDHP NIVKLFQVIE TEKTLYLIME YASGGEVFDY LVLHGRMKEK EARVKFRQIV 360
361 SAVQYCHQKR IIHRDLKAEN LLLDSELNIK IADFGFSNEF TPGSKLDTFC GSPPYAAPEL 420
421 FQGKKYDGPE VDVWSLGVIL YTLVSGSLPF DGSTLRELRE RVLRGKYRIP FYMSTDCENL 480
481 LRKFLVLNPA KRASLETIMG DKWMNMGFEE DELKPYIEPK ADLADPKRIE ALVAMGYNRS 540
541 EIEASLSQVR YDDVFATYLL LGRKSTDPES DGSRSGSSLS LRNISGNDAG ANAGSASVQS 600
601 PTHRGVHRSI SASSTKPSRR ASSGVGPTNA AATVAAATGA VGAVNPSNNY NAAGSAADRA 660
661 SVGSNFKRQN TIDSATIKEN TARLAAQNQR PASATQKMLT TADTTLNSPA KPRTATKYDP 720
721 TNGNRTVSGT SGIIPRRSTT LYEKTSSTEK TNVIPAETKA RNNTALEYSG TSGASGDSSH 780
781 PGRMSFFSKL SSRFSKRPTI ADEAAKPRVL RFTWSMKTTS PLMPDQIMQK IREVLDQNNC 840
841 DYEQRERFVL WCVHGDPNTD SLVQWEIEVC KLPRLSLNGV RFKRISGTSI GFKNIASRIA 900
901 FDLKL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
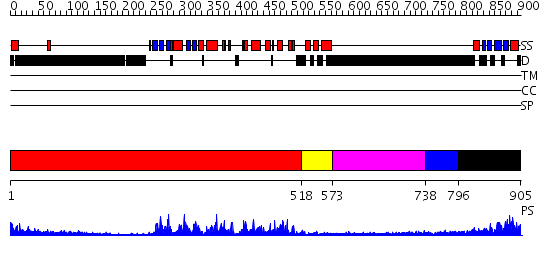
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..517] | 65.69897 | No description for 2h8hA was found. | |
| 2 | View Details | [518..572] | 97.221849 | No description for 2qnjA was found. | |
| 3 | View Details | [573..737] | 1.01 | No description for 2j63A was found. | |
| 4 | View Details | [738..795] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [796..905] | 37.0 | Solution structure of kinase associated domain 1 of mouse MAP/microtubule affinity-regulating kinase 3 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)