













| General Information: |
|
| Name(s) found: |
pita-PB /
FBpp0072022
[FlyBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 623 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
DNA replication
[IMP]
DNA endoreduplication [IMP] mitosis [NAS] |
| Molecular Function: |
DNA binding
[IDA]
zinc ion binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPGHICLECR LLFEHCYRFK QMCKRAETLL RQYPLTGNWP SPLEKPRAPM TMVASKKLLV 60
61 VPAKTAEPSE TPKKLLNTMA KSSSQVIIED VQVLESAMVT PRTVAGSSPV PRRSHAYELK 120
121 VDNNQELSMD DVQSMLEDMA SELEKEFPDI PQKASPVKPK VLNKSSIRIL NKGPAAPVEP 180
181 RLATPKVKRD DSGNVAIVTE VLDSDLPLDD QDDPTKNAEK VATDVFPCPD CERSFPLQQL 240
241 LEIHRLNHTR SRSFQCLLCE KSFFSKYDLA KHNFVHTGER PFKCAICSKA FTRKALLHRH 300
301 ERTHTDVPKF ICVYCEKPFL SRQEMEKHAE RHQKKRPFQC GVCTKSFAFK QGLERHETVH 360
361 STNLPFPCQH CERSFSTASK LARHLVAHAG KRAYPCKYCH KSYMLSHHLS RHLRTHTQTS 420
421 DASFVCSECK VSYSNYNDLL DHALIHATAS LKCPMCRKQI EDIDSVESHM DQHKQSERHA 480
481 CEFCDHIFLT QKCLQRHIED DHVVEMEPYQ NEFEDDGEGE GGVDEKEEHL DDFEDMDNVK 540
541 QEEFVTEYLE DDALYEDHLD DSDESFTPPP PKQRKTNPPK DAQQSVRQTR SRDAQRITQK 600
601 TGKNEGKPHH KLERNLKNRR SAK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
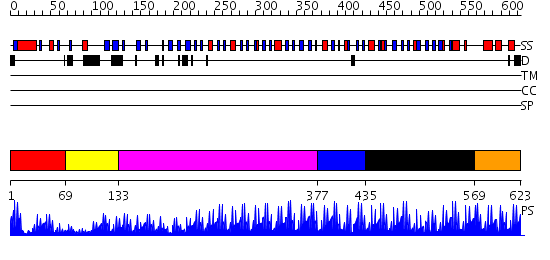
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..68] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [69..132] | 1.297996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [133..376] | 62.221849 | No description for 2i13A was found. | |
| 4 | View Details | [377..434] | 4.78 | TATA box-binding zinc finger, TATAZF | |
| 5 | View Details | [435..568] | 20.154902 | Transcription factor IIIA, TFIIIA | |
| 6 | View Details | [569..623] | 20.39794 | Ying-yang 1 (yy1, zinc finger domain) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)