













| General Information: |
|
| Name(s) found: |
gi|85816258
[NCBI NR]
gi|84796203 [NCBI NR] gi|73621137 [NCBI NR] |
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1403 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA]
mitochondrion [IDA] cytosol [IDA] |
| Biological Process: |
mitochondrion inheritance
[IMP]
spindle assembly [IMP] centrosome separation [IMP] mitotic chromosome condensation [IMP] autophagic cell death [IEP] male meiosis [IMP] spindle assembly involved in male meiosis [IMP] syncytial blastoderm mitotic cell cycle [IMP] salivary gland cell autophagic cell death [IEP] |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSAKQSGSPK AGTSPLDKRV LQPKAQQQQQ QTKQTASQNN NAQNTASNKK TQQQQQPQQG 60
61 ATTTTQTTTA AAAAATAAAA ATAATTSAAA ATSSAAAVPA ATTETATTTT VDASQADALA 120
121 KVVVKDKKKV NQKASDFSNV GDWPTLIGGT SGSGKATSNE PKRNPTKKQQ SAKTAAAVAA 180
181 SSNTTSSEVA TESNVAGTTS SSNSNPSSST TTTNTTTNSQ ATTAPVASTS HDAKAQRDAE 240
241 PAANSAAGTT GTAAGPALTK KIPKHKWRPL PIDLAKSSRP KPIGGRPNRR FSDDIADQRR 300
301 PPRVYHDRQP GAGGAGVESR HPHAGRHPYG SRPATATSER VDSWRSSSST TTAAFDEQRS 360
361 GAAGGAGAAG TGVGSATRGP RRYRTPYRGG RQGRGGFSRQ GPGRPTYRIP RHLLASGEYA 420
421 NYLPADAAGA DSQSSYVLMG THYFGNVPAA YIELDANSIK EAIKKQVEYY FSVDNLTGDF 480
481 FLRRKMDPEG YIPVTLIASF HRVLALTTDV AVIVNAIKES DKLELFEGYK VRTKTTPTTW 540
541 PITEVPEVNE GEPKAIGTLE QEQLEQNDGQ EKLEEQTEAD SPPPILTSAM ATKPLNSIPP 600
601 PPMPRNPQNL VPKMLQDKQQ SRSSTIAALN SVNAISALTQ QVEGGAAELA GHLSGLAESV 660
661 KPKSTSTPDK RNAASAGNGA GSAAALVAEP EGIWKEVKRR SKTNAIKENA TTPPQQQQPP 720
721 LSQTLNNNND NVKTNNTSSK SKSSSNNAPS NASSSATVCV TTNNASSATK ATTKTTTTST 780
781 ATTTTNNNIK SGNAAYSKTH SKSSSKTAAP PSAQCHAEKE ELDFQFDEEL MDPLPPGTGR 840
841 INNFTENFSD DDESDYEFAD RDINKLLIVA QVGRAPKHEG YDRTADFTSR TKITQDLENI 900
901 INDGLVNYEE DLWTTTNVVA DYKTVNVISQ ADFEKLAGGR NKSVLPPQVV PPPPPFEEDL 960
961 DETLVGDTTL NSTLNNTLKS RRARFYAAPN SHSIDPRTPR KRKLRHTANP PVEAHVGWLL1020
1021 DTVEHRPRTT SMGSSAGTSP TASSYGSFGS SVPQSLPVFQ HPSHALLKEN NFTQQAYHKY1080
1081 HSRCLKERRR LGYGQSQEMN TLYRFWSFFL RENFNKSMYN EFRSLALEDA GNGFRYGLEC1140
1141 LFRFFSYGLE KKFRPNIYQD FQDETIADYE TGQLYGLEKF WAFLKYYKNG EKLEVQPKLA1200
1201 EYLKSFKNIE DFRVVEPEIN EMLQGVGSLN PGRQLNRHRS VSESDGTAVI AAGGRRLNTT1260
1261 ITNRSDYVGR LLQQQHQQQQ QQQHHQYQQG YGGYNQQQNR RRTGSFGSTT VRIRSGSLGN1320
1321 KPQVANRNQG SQHELRRGGS NSGLAPHKRQ QQQKPKPGAG SQTGSTRATT SAAATATTAA1380
1381 SAATSTAATP AVTVSSGSSS SKK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
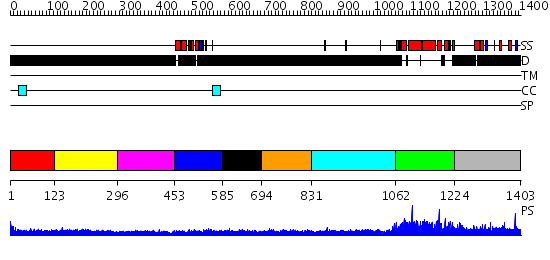
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..122] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [123..295] | 2.194995 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [296..452] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [453..584] | 2.17 | Solution structure of the La domain of c-Mpl binding protein | |
| 5 | View Details | [585..693] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [694..830] | 2.191984 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [831..1061] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1062..1223] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1224..1403] | 1.027967 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 |
|
|||||||||||||||
| 5 | No functions predicted. | |||||||||||||||
| 6 | No functions predicted. | |||||||||||||||
| 7 | No functions predicted. | |||||||||||||||
| 8 | No functions predicted. | |||||||||||||||
| 9 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)