













| General Information: |
|
| Name(s) found: |
gi|4191610
[NCBI NR]
gi|27552766 [NCBI NR] gi|18204201 [NCBI NR] |
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 598 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNKLYIGNLS PAVTADDLRQ LFGDRKLPLA GQVLLKSGYA FVDYPDQNWA IRAIETLSGK 60
61 VELHGKIMEV DYSVSKKLRS RKIQIRNIPP HLQWEVLDGL LAQYGTVENV EQVNTDTETA 120
121 VVNVTYATRE EAKIAMEKLS GHQFENYSFK ISYIPDEEVS SPSPPQRAQR GDHSSREQGH 180
181 APGGTSQARQ IDFPLRILVP TQFVGAIIGK EGLTIKNITK QTQSRVDIHR KENSGAAEKP 240
241 VTIHATPEGT SEACRMILEI MQKEADETKL AEEIPLKILA HNGLVGRLIG KEGRNLKKIE 300
301 HETGTKITIS SLQDLSIYNP ERTITVKGTV EACASAEIEI MKKLREAFEN DMLAVNQQAN 360
361 LIPGLNLSAL GIFSTGLSVL SPPAGPRGAP PAAPYHPFTT HSGYFSSLYP HHQFGPFPHH 420
421 HSYPEQEIVN LFIPTQAVGA IIGKKGAHIK QLARFAGASI KIAPAEGPDV SERMVIITGP 480
481 PEAQFKAQGR IFGKLKEENF FNPKEEVKLE AHIRVPSSTA GRVIGKGGKT VNELQNLTSA 540
541 EVIVPRDQTP DENEEVIVRI IGHFFASQTA QRKIREIVQQ VKQQEQKYPQ GVASQRSK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
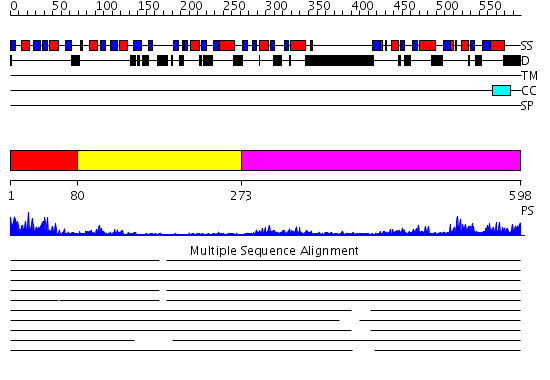
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..79] | 10.522879 | Solution structure of the RNA binding domain of IGF-II mRNA-binding protein 2 | |
| 2 | View Details | [80..272] | 4.16 | Solution structure of Polypyrimidine Tract Binding protein RBD34 complexed with CUCUCU RNA | |
| 3 | View Details | [273..598] | 34.09691 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)