













| General Information: |
|
| Name(s) found: |
gi|62472797
[NCBI NR]
gi|62472788 [NCBI NR] gi|61679377 [NCBI NR] gi|61679376 [NCBI NR] gi|51338797 [NCBI NR] gi|45549229 [NCBI NR] gi|45446625 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 1256 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endocytic vesicle
[IDA]
myosin complex [ISS][NAS] germline ring canal [IDA] cytoplasm [IDA][NAS] basal part of cell [IDA] cell cortex [IDA] unconventional myosin complex [ISS][NAS] sperm individualization complex [IDA] myosin VI complex [IPI] filamentous actin [IDA] microtubule associated complex [IDA] investment cone [IDA] |
| Biological Process: |
dorsal closure
[IEP][IMP]
spindle organization [IMP] sperm individualization [IGI][IMP] ovarian follicle cell migration [IMP] larval chitin-based cuticle development [IMP] actin filament organization [IMP] border follicle cell migration [IMP] metamorphosis [IMP] basal protein localization [IGI][IMP][TAS] actin filament-based movement [IDA] morphogenesis of follicular epithelium [IMP] nucleus localization [IMP] pseudocleavage during syncytial blastoderm formation [IMP][TAS] cytoskeleton-dependent cytoplasmic transport, nurse cell to oocyte [IDA][IMP] imaginal disc morphogenesis [IMP] establishment of mitotic spindle localization [IMP] spermatid development [IMP] regulation of protein localization [IMP] spermatogenesis [IMP] nucleus organization [IMP] cell-cell junction maintenance [TAS] sperm motility [IMP] regulation of actin cytoskeleton organization [IMP] asymmetric protein localization involved in cell fate determination [IMP][TAS] oogenesis [NAS] neuroblast division [IMP] cellular membrane organization [TAS] asymmetric neuroblast division [TAS] regulation of actin filament-based process [IGI][IMP] actin cytoskeleton organization [IMP] |
| Molecular Function: |
actin filament binding
[IDA]
ATP binding [IEA] calmodulin binding [IPI] protein binding [IPI] actin binding [IDA][IMP][NAS] myosin VI light chain binding [IPI] myosin light chain binding [IPI] ATPase activity, coupled [ISS][NAS] microtubule binding [IDA] motor activity [IEA][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRKMLEDTQL VWVRDAAEGY IQGRITEIGA KEFEVTPTDR KYPKRTCHFD DIHSSCDGPQ 60
61 DHDDNCELML LNEATFLDNL KTRYYKDKIY TYVANILIAV NPYREIKELY APDTIKKYNG 120
121 RSLGELPPHV FAIADKAIRD MRVYKLSQSI IVSGESGAGK TESTKYLLKY LCYSHDSAGP 180
181 IETKILDANP VLEAFGNAKT TRNNNSSRFG KFIEVHYDAK CQVVGGYISH YLLEKSRICT 240
241 QSAEERNYHV FYMLLAGAPQ QLRDKLSLGK PDDYRYLSGC TQYFANAKTE QLIPGSQKSK 300
301 NHQQKGPLKD PIIDDYQHFH NLDKALGRLG LSDTEKLGIY SLVAAVLHLG NIAFEEIPDD 360
361 VRGGCQVSEA SEQSLTITSG LLGVDQTELR TALVSRVMQS KGGGFKGTVI MVPLKIYEAS 420
421 NARDALAKAI YSRLFDRIVG LINQSIPFQA SNFYIGVLDI AGFEYFTVNS FEQFCINYCN 480
481 EKLQKFFNDN ILKNEQELYK REGLNVPEIT FTDNQDIIEL IEAKSNGIFT LLDEESKLPK 540
541 PSYSHFTAEV HKSWANHYRL GLPRSSRLKA HRTLRDEEGF LVRHFAGAVC YNTEQFIEKN 600
601 NDALHASLEG LVQECDNPLL QTLFPSGSST SVRGKLNFIS VGSKFKTQLG ELMEKLEQNG 660
661 TNFIRCIKPN SKMIDRQFEG SLALAQLKCS GTISVLELME HGYPSRVLFA DLYSMYKSVL 720
721 PPELVSLPAR TFCEAMFQSL NLSAKDFKFG ITKVFFRPGK FVEFDRIMRS DPENMLAIVA 780
781 KVKKWLIRSR WVKSALGALC VIKLRNRIIY RNKCVLIAQR IARGFLARKQ HRPRYQGIGK 840
841 INKIRTNTLK TIEIASGLKM GREEIISGVN DIYRQIDDAI KKIKMNPRIT QREMDSMYTV 900
901 VMANMNKLTV DLNTKLKEQQ QAEEQERLRK IQEALEAERA AKEAEEQRQR EEIENKRLKA 960
961 EMETRRKAAE AQRLRQEEED RRAALALQEQ LEKEAKDDAK YRQQLEQERR DHELALRLAN1020
1021 ESNGQVEDSP PVIRNGVNDA SPMGPNKLIR SENVRAQQQA LGKQKYDLSK WKYSELRDAI1080
1081 NTSCDIELLE ACRQEFHRRL KVYHAWKAKN RKRTTMDENE RAPRSVMEAA FKQPPLVQPI1140
1141 QEIVTAQHRY FRIPFMRANA PDNTKRGLWY AHFDGQWIAR QMELHADKPP ILLVAGTDDM1200
1201 QMCELSLEET GLTRKRGAEI LEHEFNREWE RNGGKAYKNL GAAKPNGPAA AMQKQQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
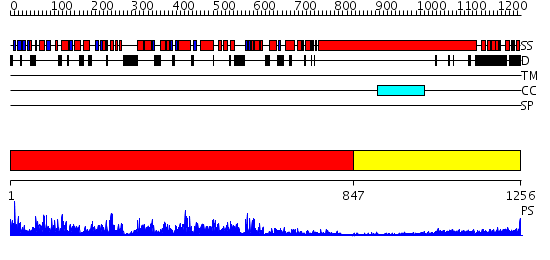
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..846] | 1000.0 | Myosin VI nucleotide-free (MDinsert2-IQ) crystal structure. | |
| 2 | View Details | [847..1256] | 9.0 | No description for 2i1jA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)