













| General Information: |
|
| Name(s) found: |
ATG4_YEAS7
[Swiss-Prot]
|
| Description(s) found:
Found 3 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae YJM789 |
| Length: | 494 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQRWLQLWKM DLVQKVSHGV FEGSSEEPAA LMNHDYIVLG EVYPERDEES GAEQCEQDCR 60
61 YRGEAVSDGF LSSLFGREIS SYTKEFLLDV QSRVNFTYRT RFVPIARAPD GPSPLSLNLL 120
121 VRTNPISTIE DYIANPDCFN TDIGWGCMIR TGQSLLGNAL QILHLGRDFR VNGNESLERE 180
181 SKFVNWFNDT PEAPFSLHNF VSAGTELSDK RPGEWFGPAA TARSIQSLIY GFPECGIDDC 240
241 IVSVSSGDIY ENEVEKVFAE NPNSRILFLL GVKLGINAVN ESYRESICGI LSSTQSVGIA 300
301 GGRPSSSLYF FGYQGNEFLH FDPHIPQPAV EDSFVESCHT SKFGKLQLSE MDPSMLIGIL 360
361 IKGEKDWQQW KLEVAESAII NVLAKRMDDF DVSCSMDDVE SVSSNSMKKD ASNNENLGVL 420
421 EGDYVDIGAI FPHTTNTEDV DEYDCFQDIH CKKQKIVVMG NTHTVNANLT DYEVEGVLVE 480
481 KETVGIHSPI DEKC |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
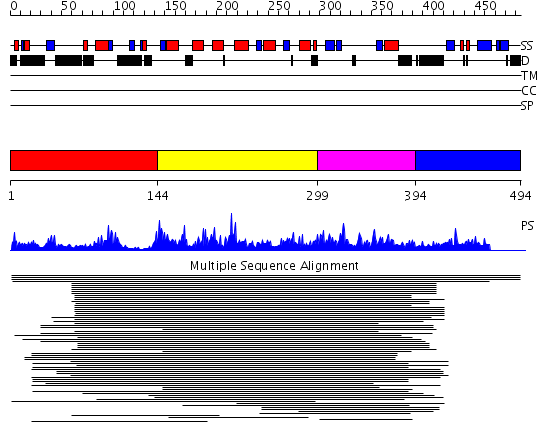
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..143] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [144..298] | 1.035968 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [299..393] | 2.035971 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [394..494] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)