













| General Information: |
|
| Name(s) found: |
gi|151943567
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae YJM789 |
| Length: | 588 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTLSDSDTE TEVVSRNLCG IVDIGSNGIR FSISSKAAHH ARIMPCVFKD RVGLSLYEVQ 60
61 YNTHTNAKCP IPRDIIKEVC SAMKRFKLIC DDFGVPETSV RVIATEATRD AINADEFVNA 120
121 VYGSTGWKVE ILGQEDETRV GIYGVVSSFN TVRGLYLDVA GGSTQLSWVI SSHGEVKQSS 180
181 KPVSLPYGAG TLLRRMRTDD NRALFYEIKE AYKDAIEKIG IPQEMIDDAK KEGGFDLWTR 240
241 GGGLRGMGHL LLYQSEGYPI QTIINGYACT YEEFSSMSDY LFLKQKIPGS SKEHKIFKVS 300
301 DRRALQLPAV GLFMSAVFEA IPQIKAVHFS EGGVREGSLY SLLPKEIRAQ DPLLIASRPY 360
361 APLLTEKYLY LLRTSIPQED IPEIVNERIA PALCNLAFVH ASYPKELQPT AALHVATRGI 420
421 IAGCHGLSHR ARALIGIALC SRWGGNIPES EEKYSQELEQ VVLREGDKAE ALRIVWWTKY 480
481 IGTIMYVICG VHPGGNIRDN VFDFHVSKRS EVETSLKELI IDDANTTKVK EESTRKNRGY 540
541 EVVVRISKDD LKTSASVRSR IITLQKKVRK LSRGSVERVK IGVQFYEE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
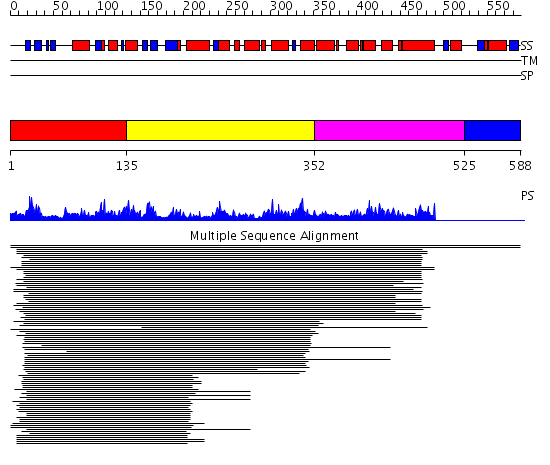
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..134] | 10.23 | Prokaryotic actin homolog MreB | |
| 2 | View Details | [135..351] | 10.23 | Prokaryotic actin homolog MreB | |
| 3 | View Details | [352..524] | 1.040981 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [525..588] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)