













| General Information: |
|
| Name(s) found: |
gi|151945402
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae YJM789 |
| Length: | 818 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAFSRLTSTH QSNHNGYSNS NKKGQSLPLT LSIDVESPPC VLYGSAMESS GAVLSGLFTV 60
61 TVVDPYSSAE DKSLKNTESN VSTTSKSLKR KSTFGSALSS RLSSLSASTS NISPSTSSTS 120
121 ISHSPTPANL RIMAGYTKIT ITSVTLSLVQ KIHFHKPFVP NISSMQTCMN CKTKITNMKS 180
181 WEIQSNTQDL SVGSHSYPFS YLIPGSVPCS SSLGATAETQ VKYELIAVVT YIDPHRNSFS 240
241 SGHSTPRKEG SSSKKRLLQL AMPIAVTRSI PRGPDKNSLR VFPPTELTAA AVLPNVVYPK 300
301 STFPLEMKLD GVSSGDRRWR MRKLSWRIEE TTRVKAHACP VHKHELRQLE EQVKIKESEK 360
361 SKKPRSHIKR YGELGPQIRV AVNSLENMPS QRLPGEPGRE QAPNSSGPAS TGNVGLDDEN 420
421 PVNEDEEDQP GSEFIHPSDD ALRQELLMQQ QRARQQQLQQ ELKNNSSLFT EEVRIISKGE 480
481 MKSGWKTDFD NNGKIELVTE IDCMGLNSGV SNPVMHASTL QTPSTGNKKP SINVACDIQD 540
541 PNLGLYVSHI LAVEIVVAEE TLQYANGQPI RKPNSKNKKE TNNNTMNVHN PDQRLAELSP 600
601 IFANRNTPKV RRMGPEDITP VNSNKSNHST NKEKASNGAS NSNIVSVPTG AARVLRMQFR 660
661 LTVTERSGLG ISWDEEVPPI YQDVELLSPP CYELSINNGI KNKLYSTMST PVRSEDDFVG 720
721 GSDEDIGNYE SQGLEPGPNV QEVTITQNKL TIPPTAHHYQ PASSSQRSLT TVQSPPLESV 780
781 VSVQGSVPFR GHVLTPHSTR DIRIQNFSDF LDSNRITQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
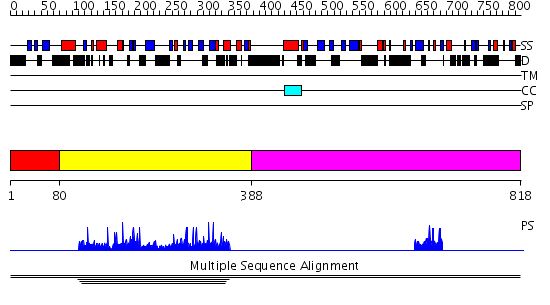
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..79] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [80..387] | 3.004923 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [388..818] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)