













| General Information: |
|
| Name(s) found: |
gcy-36 /
CE39007
[WormBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 675 amino acids |
Gene Ontology: |
|
| Cellular Component: |
soluble fraction
[ISS |
| Biological Process: |
cGMP biosynthetic process
[IEA feeding behavior [IMP] cyclic nucleotide biosynthetic process [IEA |
| Molecular Function: |
phosphorus-oxygen lyase activity
[IEA guanylate cyclase activity [IEA heme binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFGFIHESIR QLMIRTYGEA FWSKVLERAG FEAGKENIIN HYYSDADTFS LVDAVSVILK 60
61 VTREQVWEMY GCFLIQYTME TGWDDLIRSM SPNLKGFLDN LDSLHYFIDH VVYKANLRGP 120
121 SFRCEDNPDG TITLHYYTGR PGLYPIVKGV LREAAKRVFK LDVSMTITGR TQRSVQMATG 180
181 ERIEEHVIFL VKTLNTDQSN EEALGTAVVQ HSNNYKIRLT HMDFISTFPY HMVVDQDCKI 240
241 VQVGRELYNH IPKDLLSVGT PLMRIFEVTR PQIPLDFDSI CNFINAVFVL QVKTTPMEFQ 300
301 RNANKRAAQA IEASENLYED NNGALALSQS QHLKLKGQMM LMSSGGHIMY LCSPYVTSIP 360
361 ELLQYGLRLT AMPIHDPTRD LILLNQQRLS DVEMNLQLEA NNEQLENMAK DLEVEKGKTD 420
421 ALLREMLPPS VAQQLKQGLS VEAREYEEAT VMFTDVPTFQ QIVPLCTPKD IVHLLNELFT 480
481 KFDRLIGIQK AYKVETVGDS YMSVGGIPDL VDDHCEVICH LALGMVMEAR TVCDPITNTP 540
541 LHIRAGIHSG PVVAGVVGAK MPRYCLFGDT VNTSSRMESH SPIGRIHCSE NAKKCAESTG 600
601 RFEFEPRGRV QIKGKGEMNT YFLLRSFKRS IWEIIDRRRD ENCNSIDGYN ELREGYVDDV 660
661 LNKVTQKNSK TCSIS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
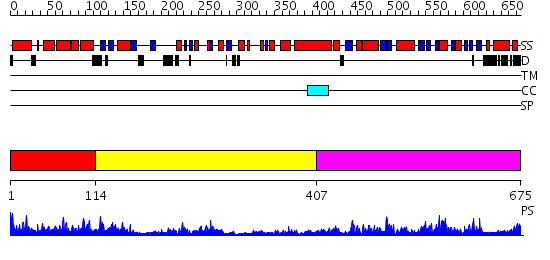
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..113] | 43.09691 | No description for 2o09A was found. | |
| 2 | View Details | [114..406] | 7.0 | Musk tyrosine kinase | |
| 3 | View Details | [407..675] | 49.522879 | Crystal structure of a mutant form of the mycobacterial adenylyl cyclase Rv1625c |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)