













| General Information: |
|
| Name(s) found: |
gi|30725460
[NCBI NR]
gi|25405328 [NCBI NR] gi|22330139 [NCBI NR] gi|20259988 [NCBI NR] gi|18252161 [NCBI NR] gi|17065304 [NCBI NR] gi|10120437 [NCBI NR] |
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 629 amino acids |
Gene Ontology: |
|
| Cellular Component: |
peroxisome
[IDA]
mitochondrion [IDA] |
| Biological Process: |
response to oxidative stress
[IEP][IGI]
metabolic process [ISS] |
| Molecular Function: |
zinc ion binding
[IEA][ISS]
oxidoreductase activity [IEA][ISS] catalytic activity [IEA] binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEIKPGLSAL VTGGASGIGR ALCLALAEKG VFVTVADFSE EKGQETTSLV REANAKFHQG 60
61 LSFPSAIFVK CDVTNRGDLL AAFDKHLATF GTLDICINNA GISTPLRFDK DDTDGSKSWK 120
121 HTINVDLIAV VEGTQLAIKA MKAKQKPGVI INMGSAAGLY PMPVDPIYAA SKAGVVLFTR 180
181 SLAYYRRQGI RINVLCPEFI KTDLAEAIDA SILESIGGYM SMDMLIKGAF ELITDEKKAG 240
241 ACLWITKRRG LEYWPTPMEE TKYLVGSSSR KRPSFKVSTK IEFPQSFEKM IVHTLSHKFR 300
301 SATRIVRAPL QLPIGPHQVL LKIIYAGVNA SDVNFSSGRY FTGGSPKLPF DAGFEGVGLI 360
361 AAVGESVKNL EVGTPAAVMT FGAYSEYMIV SSKHVLPVPR PDPEVVAMLT SGLTALIALE 420
421 KAGQMKSGET VLVTAAAGGT GQFAVQLAKL SGNKVIATCG GSEKAKLLKE LGVDRVIDYK 480
481 SENIKTVLKK EFPKGVNIIY ESVGGQMFDM CLNALAVYGR LIVIGMISQY QGEKGWEPAK 540
541 YPGLCEKILA KSQTVAGFFL VQYSQLWKQN LDKLFNLYAL GKLKVGIDQK KFIGLNAVAD 600
601 AVEYLHSGKS TGKVVVCIDP AFEQKTSRL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
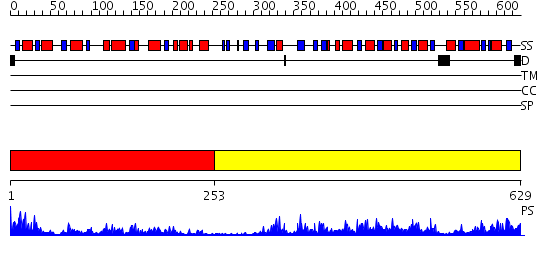
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..252] | 33.39794 | No description for 2gdzA was found. | |
| 2 | View Details | [253..629] | 67.522879 | Structure of the MGC45594 gene product |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|