













| General Information: |
|
| Name(s) found: |
gi|9294249
[NCBI NR]
gi|24030319 [NCBI NR] gi|22316560 [NCBI NR] gi|15232528 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 690 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA]
spindle [IDA] |
| Biological Process: |
protein amino acid phosphorylation
[IEA]
|
| Molecular Function: |
ATP binding
[IEA]
protein kinase activity [IEA] protein serine/threonine kinase activity [IEA] kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDDVAGLQEA AGARFSQIEL IGRGSFGDVY KAFDKDLNKE VAIKVIDLEE SEDEIEDIQK 60
61 EISVLSQCRC PYITEYYGSY LHQTKLWIIM EYMAGGSVAD LLQSNNPLDE TSIACITRDL 120
121 LHAVEYLHNE GKIHRDIKAA NILLSENGDV KVADFGVSAQ LTRTISRRKT FVGTPFWMAP 180
181 EVIQNSEGYN EKADIWSLGI TVIEMAKGEP PLADLHPMRV LFIIPRETPP QLDEHFSRQV 240
241 KEFVSLCLKK APAERPSAKE LIKHRFIKNA RKSPKLLERI RERPKYQVKE DEETPRNGAK 300
301 APVESSGTVR IARDERSQGA PGYSFQGNTV KNAGWDFTVG GSQSIGTVRA LKPPQARERR 360
361 QEVSPNRISQ RTTRPSGNQW SSATGSTISE ASEGGFVRRH PFQNDHEDGF HEEDDSSLSG 420
421 SGTVVIRTPR SSQSSSVFRE PSSGSSGRYA AFDDASASGT VVVRGQYDDS GSPRTPKSRL 480
481 GIQERTSSAS EDSNANLAEA KAALDAGFRR GKARERLGMG NNNNDGKVNR RREQMADDSD 540
541 YSRNSGDKSS KQKVVPRSEQ VSDEEDDSIW ESLPASLSVL LIPSLKEALG DDSKESTVRT 600
601 VSRSLVMMER EKPGSCEAFV AKLIELLGSS KEASVKELHD MAVCVFAKTT PDNAENKMKQ 660
661 ANKEFSSNTN VSPLGRFLLS RWLGQSSRDL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
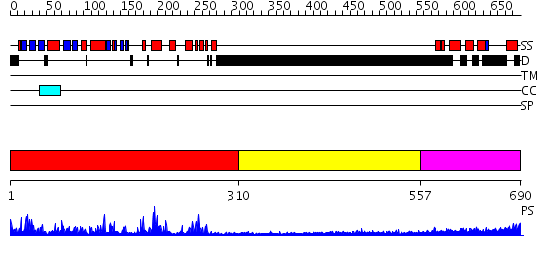
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..309] | 55.0 | Crystal Structure of the TAO2 Kinase Domain: Activation and Specifity of a Ste20p MAP3K | |
| 2 | View Details | [310..556] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [557..690] | 3.221849 | Heavy meromyosin subfragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)