













| General Information: |
|
| Name(s) found: |
CHR19_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 763 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IEA]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
nucleic acid binding
[IEA]
DNA binding [IEA][ISS] ATP binding [IEA][ISS] helicase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKRDFDEISE EEWSQHSFNA SRVLKRPRTP KKTRAATNPT PSIESFAFRR PSTAMTIESN 60
61 SSDGDCVEIE DLGDSDSDVK IVNGEDLLLE DEEEVEETKV VMRAARVGRR FVIEDEEASD 120
121 DDDDEAESSA SEDEFGGGGG GSGGRRGEDE DVVGKALQKC AKISADLRKE LYGTSSGVTD 180
181 RYSEVETSTV RIVTQNDIDD ACKAEDSDFQ PILKPYQLVG VNFLLLLYKK GIEGAILADE 240
241 MGLGKTIQAI TYLTLLSRLN NDPGPHLVVC PASVLENWER ELRKWCPSFT VLQYHGAARA 300
301 AYSRELNSLS KAGKPPPFNV LLVCYSLFER HSEQQKDDRK VLKRWRWSCV LMDEAHALKD 360
361 KNSYRWKNLM SVARNANQRL MLTGTPLQND LHELWSLLEF MLPDIFTTEN VDLKKLLNAE 420
421 DTELITRMKS ILGPFILRRL KSDVMQQLVP KIQRVEYVLM ERKQEDAYKE AIEEYRAASQ 480
481 ARLVKLSSKS LNSLAKALPK RQISNYFTQF RKIANHPLLI RRIYSDEDVI RIARKLHPIG 540
541 AFGFECSLDR VIEEVKGFND FRIHQLLFQY GVNDTKGTLS DKHVMLSAKC RTLAELLPSM 600
601 KKSGHRVLIF SQWTSMLDIL EWTLDVIGVT YRRLDGSTQV TDRQTIVDTF NNDKSIFACL 660
661 LSTRAGGQGL NLTGADTVII HDMDFNPQID RQAEDRCHRI GQTKPVTIFR LVTKSTVDEN 720
721 IYEIAKRKLV LDAAVLESGV HVDDNGDTPE KTMGEILASL LMG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
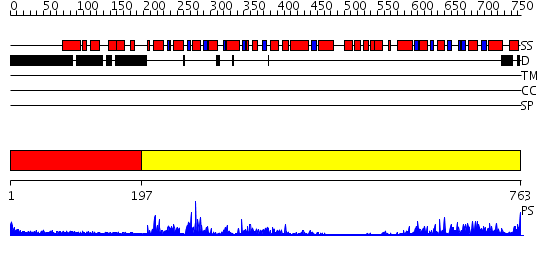
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..196] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [197..763] | 78.522879 | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)