













| General Information: |
|
| Name(s) found: |
gi|7270668
[NCBI NR]
gi|71143064 [NCBI NR] gi|4006861 [NCBI NR] gi|25407796 [NCBI NR] gi|194306666 [NCBI NR] gi|15235474 [NCBI NR] |
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 562 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA]
plasma membrane [IDA] |
| Biological Process: |
protein polymerization
[IEA]
|
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MREIVTIQVG EFANFVGSHF WNFQDELLGL ASDPESDPIF RNHNLDMDVL YRSGETQQGV 60
61 ATYTPRLVSV NLKGALGTMS SRGTLYNEGS SSRSDSSATW FGDVDTQRSE PRKRNLFLQS 120
121 LYEEEHVVGK EKAKEIEDKD IVGCLDEEVE CWTDFSKSHY HPQSLYELNG LWMDSQAFNN 180
181 YGIGKDVFSE ASRGEEICDR LRFFVEECDH IQGIKFLVDD SGGFSAVAAD FLENMADEYT 240
241 NVPVLLYSVR TPMSQMSSKK TVSNKLHDAI SFSRLSSFCK LFTPIGLPSL TGSKASKFLN 300
301 LGDEKPYRSS AVYAAALHSS TIPFRMQPTS SDSSEVSNSM DVNTLVQLLT GRGRQNIVAI 360
361 LDSAMPAPTL AGKQLENTLL TSLQALTPEV TEDVEDNQAV ESMCILGALR SEDKEALVSE 420
421 VKNAVDASYE QATTKGKPLF CNLSVSRCPL PVPLPFPSIF GNLVGRKGEI LSSPVSDSLY 480
481 RGSLDVHSIP VATRWRSSSA ILPFLETRMG NLEKLGIQWG AMGSDVVRAW GFGREELQEM 540
541 RENLSKMVSE LNPQFFESSD SD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
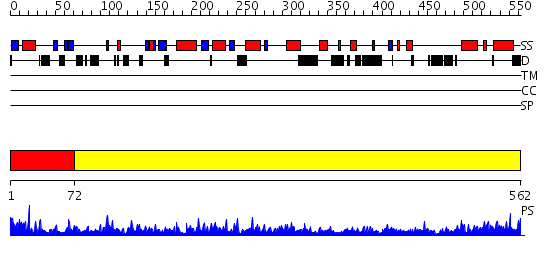
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..71] | 3.47 | Structure of BtubA from Prosthecobacter dejongeii | |
| 2 | View Details | [72..562] | 145.0 | Tubulin-colchicine-vinblastine: stathmin-like domain complex |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)