













| General Information: |
|
| Name(s) found: |
gi|6579197
[NCBI NR]
gi|15223158 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 2271 amino acids |
Gene Ontology: |
|
| Cellular Component: |
apoplast
[IDA]
epsilon DNA polymerase complex [IPI] |
| Biological Process: |
embryonic development ending in seed dormancy
[IMP][NAS]
positive regulation of S phase of mitotic cell cycle [IMP] negative regulation of long-day photoperiodism, flowering [IMP] |
| Molecular Function: |
nucleic acid binding
[IEA]
nucleotide binding [IEA] DNA binding [IEA] zinc ion binding [IEA] DNA-directed DNA polymerase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGDNRRRDR KDTRWSKKPK VVNTAEDELE SKLGFGLFSE GETRLGWLLT FSSSSWEDRD 60
61 TGKVYSCVDL YFVTQDGFSF KTKYKFRPYF YAATKDKMEL ELEAYLRRRY ERQVADIEIV 120
121 EKEDLDLKNH LSGLQKKYLK ISFDTVQQLM EVKRDLLHIV ERNQAKFDAL EAYESILAGK 180
181 REQRPQDCLD SIVDLREYDV PYHVRFAIDN DVRSGQWYNV SISSTDVILE KRTDLLQRAE 240
241 VRVCAFDIET TKLPLKFPDA EYDQIMMISY MVDGQGFLII NREVTACVGE DVEDLEYTPK 300
301 PEFEGYFKVT NVKNEVELLQ RWFYHMQELK PGIYVTYNGD FFDWPFIERR ASHHGIKMNE 360
361 ELGFRCDQNQ GECRAKFACH LDCFAWVKRD SYLPQGSHGL KAVTKAKLGY DPLEVNPEDM 420
421 VRFAMEKPQT MASYSVSDAV ATYYLYMTYV NPFIFSLATI IPMVPDEVLR KGSGTLCEML 480
481 LMVEVSTYLP QAQAYKANVV CPNKNQADPE KFYQNQLLES ETYIGGHVEC LESGQLIDNL 540
541 GRDLEYAITV EGKMRMDSIS NYDEVKDEIK EKLEKLRDDP IREEGPLIYH LDVAAMYPNI 600
601 ILTNRLQVHL ISPPSIVTDE ICTACDFNRP GKTCLRKLEW VWRGVTFMGK KSDYYHLKKQ 660
661 IESEFVDAGA NIMSSKSFLD LPKVDQQSKL KERLKKYCQK AYKRVLDKPI TEVREAGICM 720
721 RENPFYVDTV RSFRDRRYEY KTLNKVWKGK LSEAKASGNS IKIQEAQDMV VVYDSLQLAH 780
781 KCILNSFYGY VMRKGARWYS MEMAGVVTYT GAKIIQNARL LIERIGKPLE LDTDGIWCCL 840
841 PGSFPENFTF KTIDMKKLTI SYPCVMLNVD VAKNNTNDQY QTLVDPVRKT YKSHSECSIE 900
901 FEVDGPYKAM IIPASKEEGI LIKKRYAVFN HDGTLAELKG FEIKRRGELK LIKVFQAELF 960
961 DKFLHGSTLE ECYSAVAAVA DRWLDLLDVC EISCFFWVIY LIGFLSHFLQ NQGKDIADSE1020
1021 LLDYISESST MSKSLADYGE QKSCAVTTAK RLAEFLGVTM VKDKGLRCQY IVACEPKGTP1080
1081 VSERAVPVAI FTTNPEVMKF HLRKWCKTSS DVGIRLIIDW SYYKQRLSSA IQKVITIPAA1140
1141 MQKVANPVPR VLHPDWLHKK VREKDDKFRQ RKLVDMFSSA NKDVVLDTDL PVTKDNVEDI1200
1201 EDFCKENRPS VKGPKPIARS YEVNKKQSEC EQQESWDTEF HDISFQNIDK SVNYQGWLEL1260
1261 KKRKWKVTLE KKKKRRLGDL RSSNQVDTHE INQKVGQGRG GVGSYFRRPE EALTSSHWQV1320
1321 CTIIQLVPSP QSGQFFAWVV VEGLMLKIPL SIPRVFYINS KVPIDEYFQG KCVNKILPHG1380
1381 RPCYSLTEAC VKIQEDQFKK ESKKRAALLA DPGVEGIYET KVPLEFSAIC QIGCVCKIDN1440
1441 KAKHRNTQDG WEVGELHMKT TTECHYLKRS IPLVYLYNST STGRAIYVLY CHVSKLMSAV1500
1501 VVDPFNGNEL LPSALERQFR DSCLELSLDS LSWDGIRFQV HYVDHPEAAK KIIQRAISEY1560
1561 REENCGPTVA VIECPDFTFM KEGIKALDDF PCVRIPFNDD DNSYQPVSWQ RPAAKIAMFR1620
1621 CAAAFQWLDR RITQSRYAHV PLGNFGLDWL TFTIDIFLSR ALRDQQQVLW VSDNGVPDLG1680
1681 GINNEEAFFA DEVQQTSLVF PGAYRKVSVE LKIHNLAVNA LLKSNLVNEM EGGGFMGFEQ1740
1741 DVNPRGINSN DNTSFDETTG CAQAFRVLKQ LIHSCLTDVR KSKNIYADSI LQRLSWWLCS1800
1801 PSSKLHDPAL HLMLHKVMQK VFALLLTDLR RLGAIIIYAD FSKVIIDTVK FDLSAAKAYC1860
1861 ESLLSTVRNR LVSSLLDIHV YDLKSHGLHS FSCHDFCASP LTYNYAGIRA DDEISLDEVT1920
1921 IEPKWSVARH LPEYIEVWRQ NPHSNQCSFS ENKKHVATHG PFFLDTDAFH LSWQRDFIII1980
1981 IAKFIFDPWK FAIENKKGSS ESLEAQMIEY LREQIGSTFI NMLVKKVDDI MSHMKEINVS2040
2041 DASRVSGQAP KGDYSLEFIQ VISAVLALDQ NVQQDVLLKT LFITDFIHKS NLISGNVVLE2100
2101 TMFTNRAGFF FEWVMRKSLL KYIKVKECAA EAEFLDPGPS FILPNVACSN CDAYRDLDIC2160
2161 RDPALLTEKE WSCADTQCGK IYDREQMESS LLEMVRQRER MYHMQDVVCI RCNQVKAAHL2220
2221 TEQCECSGSF RCKESGSEFS KRMEIFMDIA KRQKFRLLEE YISWIIYGPS Y |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
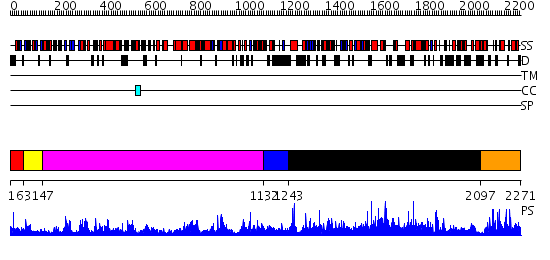
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..62] | 123.0 | No description for 2gv9A was found. | |
| 2 | View Details | [63..146] | 105.0 | Exonuclease domain of family B (archaeal and phage) DNA polymerases; T4-like DNA polymerase | |
| 3 | View Details | [147..1131] | 70.0 | CRYSTAL STRUCTURE OF ENTEROBACTERIA PHAGE RB69 GP43 DNA POLYMERASE COMPLEXED WITH 8-OXOGUANOSINE CONTAINING DNA | |
| 4 | View Details | [1132..1242] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [1243..2096] | 3.7 | Crystal structure of ESCHERICHIA coli DNA Polymerase II | |
| 6 | View Details | [2097..2271] | 2.038955 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)