













| General Information: |
|
| Name(s) found: |
gi|8843884
[NCBI NR]
gi|42570590 [NCBI NR] gi|42568255 [NCBI NR] |
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1068 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intrinsic to membrane
[IEA]
|
| Biological Process: |
signal transduction
[IEA]
innate immune response [IEA] apoptosis [IEA] defense response [ISS] |
| Molecular Function: |
nucleotide binding
[IEA]
ATP binding [IEA] transmembrane receptor activity [IEA] nucleoside-triphosphatase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALSSSLSCI KRYQVFSSFH GPDVRKGFLS HLHSVFASKG ITTFNDQKID RGQTIGPELI 60
61 QGIREARVSI VVLSKKYASS SWCLDELVEI LKCKEALGQI VMTVFYEVDP SDVKKQSGVF 120
121 GEAFEKTCQG KNEEVKIRWR NALAHVATIA GEHSLNWDNE AKMIQKIVTD VSDKLNLTPS 180
181 RDFEGMVGME AHLKRLNSLL CLESDEVKMI GIWGPAGIGK TTIARTLFNK ISSIFPFKCF 240
241 MENLKGSIKG GAEHYSKLSL QKQLLSEILK QENMKIHHLG TIKQWLHDQK VLIILDDVDD 300
301 LEQLEVLAED PSWFGSGSRI IVTTEDKNIL KAHRIQDIYH VDFPSEEEAL EILCLSAFKQ 360
361 SSIPDGFEEL ANKVAELCGN LPLGLCVVGA SLRRKSKNEW ERLLSRIESS LDKNIDNILR 420
421 IGYDRLSTED QSLFLHIACF FNNEKVDYLT ALLADRKLDV VNGFNILADR SLVRISTDGH 480
481 VVMHHYLLQK LGRRIVHEQW PNEPGKRQFL IEAEEIRDVL TKGTGTESVK GISFDTSNIE 540
541 EVSVGKGAFE GMRNLQFLRI YRDSFNSEGT LQIPEDMEYI PPVRLLHWQN YPRKSLPQRF 600
601 NPEHLVKIRM PSSKLKKLWG GIQPLPNLKS IDMSFSYSLK EIPNLSKATN LEILSLEFCK 660
661 SLVELPFSIL NLHKLEILNV ENCSMLKVIP TNINLASLER LDMTGCSELR TFPDISSNIK 720
721 KLNLGDTMIE DVPPSVGCWS RLDHLYIGSR SLKRLHVPPC ITSLVLWKSN IESIPESIIG 780
781 LTRLDWLNVN SCRKLKSILG LPSSLQDLDA NDCVSLKRVC FSFHNPIRAL SFNNCLNLDE 840
841 EARKGIIQQS VYRYICLPGK KIPEEFTHKA TGRSITIPLS PGTLSASSRF KASILILPVE 900
901 SYETDDISCS LRTKGGVEVH CCELPYHFLL RSRSEHLFIF HGDLFPQGNK YHEVDVTMSE 960
961 ITFEFSHTKI GDKIIECGVQ IMTEGAEGDS SRELDSFETE SSSSQVDNFE TGGNNNHHTD1020
1021 GNGDGNYQAE GFKFFQDENI KTSKHTGFRS WLRELGLKVK KMNSHGVS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
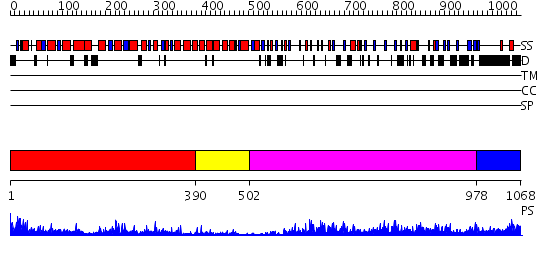
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..389] | 4.55 | Structure of the apoptotic protease-activating factor 1 bound to ADP | |
| 2 | View Details | [390..501] | 9.09691 | No description for 2z7xA was found. | |
| 3 | View Details | [502..977] | 16.30103 | No description for 2z63A was found. | |
| 4 | View Details | [978..1068] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)