













| General Information: |
|
| Name(s) found: |
VATC_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 375 amino acids |
Gene Ontology: |
|
| Cellular Component: |
vacuolar proton-transporting V-type ATPase, V1 domain
[IDA]
chloroplast [IDA] plant-type vacuole [IDA] vacuolar membrane [IDA] vacuole [IDA] plasma membrane [IDA] |
| Biological Process: |
unidimensional cell growth
[IMP]
lignin biosynthetic process [IGI] |
| Molecular Function: |
proton-transporting ATPase activity, rotational mechanism
[IGI]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTSRYWVVSL PVKDSASSLW NRLQEQISKH SFDTPVYRFN IPNLRVGTLD SLLALGDDLL 60
61 KSNSFVEGVS QKIRRQIEEL ERISGVESNA LTVDGVPVDS YLTRFVWDEA KYPTMSPLKE 120
121 VVDNIQSQVA KIEDDLKVRV AEYNNIRGQL NAINRKQSGS LAVRDLSNLV KPEDIVESEH 180
181 LVTLLAVVPK YSQKDWLACY ETLTDYVVPR SSKKLFEDNE YALYTVTLFT RVADNFRIAA 240
241 REKGFQVRDF EQSVEAQETR KQELAKLVQD QESLRSSLLQ WCYTSYGEVF SSWMHFCAVR 300
301 TFAESIMRYG LPPAFLACVL SPAVKSEKKV RSILERLCDS TNSLYWKSEE DAGAMAGLAG 360
361 DSETHPYVSF TINLA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
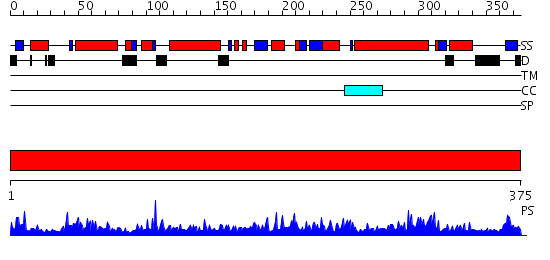
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..375] | 121.0 | Crystal Structure of subunit C (vma5p) of the yeast V-ATPase |
Functions predicted (by domain):
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)