













| General Information: |
|
| Name(s) found: |
PABP5_ARATH
[Swiss-Prot]
PAB5_ARATH [Swiss-Prot] |
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 668 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytosol [IDA] |
| Biological Process: |
translational initiation
[IGI]
nuclear-transcribed mRNA poly(A) tail shortening [IGI] |
| Molecular Function: |
translation initiation factor activity
[ISS]
RNA binding [ISS] poly(A) RNA binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVDQVIPNQP TVAAAAPPPF PAVSQVAAVA AAAAAAEALQ THPNSSLYVG DLDPSVNESH 60
61 LLDLFNQVAP VHNLRVCRDL THRSLGYAYV NFANPEDASR AMESLNYAPI RDRPIRIMLS 120
121 NRDPSTRLSG KGNVFIKNLD ASIDNKALYE TFSSFGTILS CKVAMDVVGR SKGYGFVQFE 180
181 KEETAQAAID KLNGMLLNDK QVFVGHFVRR QDRARSESGA VPSFTNVYVK NLPKEITDDE 240
241 LKKTFGKYGD ISSAVVMKDQ SGNSRSFGFV NFVSPEAAAV AVEKMNGISL GEDVLYVGRA 300
301 QKKSDREEEL RRKFEQERIS RFEKLQGSNL YLKNLDDSVN DEKLKEMFSE YGNVTSCKVM 360
361 MNSQGLSRGF GFVAYSNPEE ALLAMKEMNG KMIGRKPLYV ALAQRKEERQ AHLQSLFTQI 420
421 RSPGTMSPVP SPMSGFHHHP PGGPMSGPHH PMFIGHNGQG LVPSQPMGYG YQVQFMPGMR 480
481 PGAGPPNFMM PFPLQRQTQP GPRVGFRRGA NNMQQQFQQQ QMLQQNASRF MGGAGNRRNG 540
541 MEASAPQGII PLPLNASANS HNAPQRSHKP TPLTISKLAS DLALASPDKH PRMLGDHLYP 600
601 LVEQQEPANA AKVTGMLLEM DQAEILHLLE SPEALKAKVS EALDVLRRSA DPAAVSSVDD 660
661 QFALSSSE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
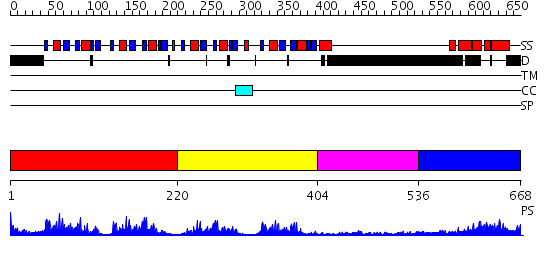
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..219] | 52.522879 | Poly(A)-binding protein | |
| 2 | View Details | [220..403] | 15.0 | Polypyrimidine tract-binding protein | |
| 3 | View Details | [404..535] | 1.22 | Crystal Structure of Rat Synapsin I C Domain Complexed to Ca.ATP | |
| 4 | View Details | [536..668] | 35.221849 | poly(A) binding protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.48 |
Source: Reynolds et al. (2008)