













| General Information: |
|
| Name(s) found: |
PABP3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 660 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
mRNA processing
[IGI]
|
| Molecular Function: |
translation initiation factor activity
[ISS]
RNA binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAAVATGVA PATMVDQVPS PTAQTSVQVP VSIPLPSPVV VADQTHPNSS LYAGDLDPKV 60
61 TEAHLFDLFK HVANVVSVRV CRDQNRRSLG YAYINFSNPN DAYRAMEALN YTPLFDRPIR 120
121 IMLSNRDPST RLSGKGNIFI KNLDASIDNK ALFETFSSFG TILSCKVAMD VTGRSKGYGF 180
181 VQFEKEESAQ AAIDKLNGML MNDKQVFVGH FIRRQERARD ENTPTPRFTN VYVKNLPKEI 240
241 GEDELRKTFG KFGVISSAVV MRDQSGNSRC FGFVNFECTE AAASAVEKMN GISLGDDVLY 300
301 VGRAQKKSER EEELRRKFEQ ERINRFEKSQ GANLYLKNLD DSVDDEKLKE MFSEYGNVTS 360
361 SKVMLNPQGM SRGFGFVAYS NPEEALRALS EMNGKMIGRK PLYIALAQRK EDRRAHLQAL 420
421 FSQIRAPGPM SGFHHPPGGP MPGPPQHMYV GQNGASMVPS QPIGYGFQPQ FMPGMRPGSG 480
481 PGNFIVPYPL QRQPQTGPRM GFRRGATNVQ QHIQQQQLMH RNPSPGMRYM NGASNGRNGM 540
541 DSSVPQGILP PIIPLPIDAS SISHQKAPLL PISKLTSSLA SASPADRTRM LGEQLYPLVE 600
601 RHEPLHVAKV TGMLLEMDQA EILHLMESPE ALKSKVSEAL DVLRLSVDPT DHDLGFSTTD 660
661 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
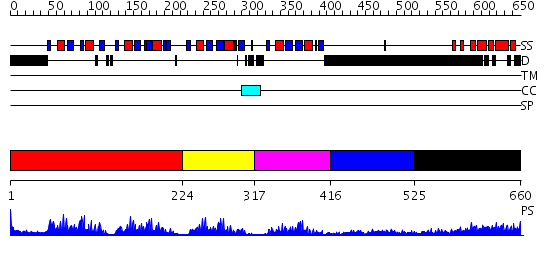
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..223] | 53.221849 | Poly(A)-binding protein | |
| 2 | View Details | [224..316] | 8.0 | Solution structure of RRM domain in TAR DNA-binding protein-43 | |
| 3 | View Details | [317..415] | 4.46 | No description for 2f3jA was found. | |
| 4 | View Details | [416..524] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [525..660] | 30.39794 | poly(A) binding protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.57 |
Source: Reynolds et al. (2008)