













| General Information: |
|
| Name(s) found: |
METE1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 765 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
peroxisome [IDA] chloroplast [IDA] membrane [IDA] apoplast [IDA] cytosol [IDA][ISS] plasma membrane [IDA] |
| Biological Process: |
methionine biosynthetic process
[ISS]
response to zinc ion [IEP] response to salt stress [IEP] response to cadmium ion [IEP] |
| Molecular Function: |
methionine synthase activity
[IDA]
copper ion binding [IDA] 5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASHIVGYPR MGPKRELKFA LESFWDGKST AEDLQKVSAD LRSSIWKQMS AAGTKFIPSN 60
61 TFAHYDQVLD TTAMLGAVPP RYGYTGGEIG LDVYFSMARG NASVPAMEMT KWFDTNYHYI 120
121 VPELGPEVNF SYASHKAVNE YKEAKALGVD TVPVLVGPVS YLLLSKAAKG VDKSFELLSL 180
181 LPKILPIYKE VITELKAAGA TWIQLDEPVL VMDLEGQKLQ AFTGAYAELE STLSGLNVLV 240
241 ETYFADIPAE AYKTLTSLKG VTAFGFDLVR GTKTLDLVKA GFPEGKYLFA GVVDGRNIWA 300
301 NDFAASLSTL QALEGIVGKD KLVVSTSCSL LHTAVDLINE TKLDDEIKSW LAFAAQKVVE 360
361 VNALAKALAG QKDEALFSAN AAALASRRSS PRVTNEGVQK AAAALKGSDH RRATNVSARL 420
421 DAQQKKLNLP ILPTTTIGSF PQTVELRRVR REYKAKKVSE EDYVKAIKEE IKKVVDLQEE 480
481 LDIDVLVHGE PERNDMVEYF GEQLSGFAFT ANGWVQSYGS RCVKPPVIYG DVSRPKAMTV 540
541 FWSAMAQSMT SRPMKGMLTG PVTILNWSFV RNDQPRHETC YQIALAIKDE VEDLEKGGIG 600
601 VIQIDEAALR EGLPLRKSEH AFYLDWAVHS FRITNCGVQD STQIHTHMCY SHFNDIIHSI 660
661 IDMDADVITI ENSRSDEKLL SVFREGVKYG AGIGPGVYDI HSPRIPSSEE IADRVNKMLA 720
721 VLEQNILWVN PDCGLKTRKY TEVKPALKNM VDAAKLIRSQ LASAK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
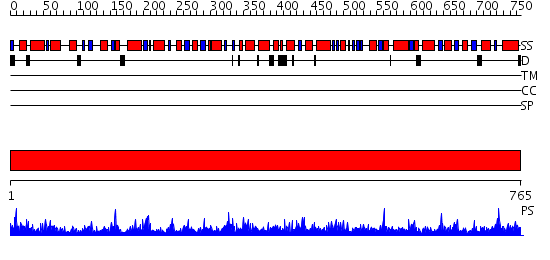
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..765] | 1000.0 | A. thaliana cobalamine independent methionine synthase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)