













| General Information: |
|
| Name(s) found: |
HSP7C_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 649 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
cytosolic ribosome [IDA] cell wall [IDA] apoplast [IDA] cytosol [TAS] vacuole [IDA] plasma membrane [IDA] |
| Biological Process: |
response to heat
[IEP]
response to cadmium ion [IEP] protein folding [TAS] |
| Molecular Function: |
ATP binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAGKGEGPAI GIDLGTTYSC VGVWQHDRVE IIANDQGNRT TPSYVAFTDS ERLIGDAAKN 60
61 QVAMNPINTV FDAKRLIGRR FTDSSVQSDI KLWPFTLKSG PAEKPMIVVN YKGEDKEFSA 120
121 EEISSMILIK MREIAEAYLG TTIKNAVVTV PAYFNDSQRQ ATKDAGVIAG LNVMRIINEP 180
181 TAAAIAYGLD KKATSVGEKN VLIFDLGGGT FDVSLLTIEE GIFEVKATAG DTHLGGEDFD 240
241 NRMVNHFVQE FKRKNKKDIS GNPRALRRLR TACERAKRTL SSTAQTTIEI DSLFDGIDFY 300
301 APITRARFEE LNIDLFRKCM EPVEKCLRDA KMDKNSIDDV VLVGGSTRIP KVQQLLVDFF 360
361 NGKELCKSIN PDEAVAYGAA VQAAILSGEG NEKVQDLLLL DVTPLSLGLE TAGGVMTVLI 420
421 QRNTTIPTKK EQVFSTYSDN QPGVLIQVYE GERARTKDNN LLGKFELSGI PPAPRGVPQI 480
481 TVCFDIDANG ILNVSAEDKT TGQKNKITIT NDKGRLSKDE IEKMVQEAEK YKSEDEEHKK 540
541 KVDAKNALEN YAYNMRNTIR DEKIGEKLAG DDKKKIEDSI EAAIEWLEAN QLAECDEFED 600
601 KMKELESICN PIIAKMYQGG EAGGPAAGGM DEDVPPSAGG AGPKIEEVD |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
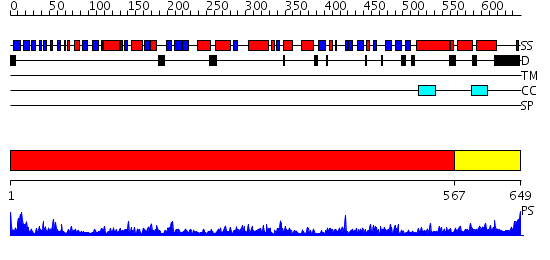
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..566] | 1000.0 | crystal structure of bovine hsc70(aa1-554)E213A/D214A mutant | |
| 2 | View Details | [567..649] | 56.30103 | DnaK |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)