













| General Information: |
|
| Name(s) found: |
CDPK1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 610 amino acids |
Gene Ontology: |
|
| Cellular Component: |
peroxisome
[IDA]
membrane [NAS] |
| Biological Process: |
protein amino acid phosphorylation
[IDA][ISS]
|
| Molecular Function: |
protein binding
[IPI]
calmodulin-dependent protein kinase activity [ISS] kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGNTCVGPSR NGFLQSVSAA MWRPRDGDDS ASMSNGDIAS EAVSGELRSR LSDEVQNKPP 60
61 EQVTMPKPGT DVETKDREIR TESKPETLEE ISLESKPETK QETKSETKPE SKPDPPAKPK 120
121 KPKHMKRVSS AGLRTESVLQ RKTENFKEFY SLGRKLGQGQ FGTTFLCVEK TTGKEFACKS 180
181 IAKRKLLTDE DVEDVRREIQ IMHHLAGHPN VISIKGAYED VVAVHLVMEC CAGGELFDRI 240
241 IQRGHYTERK AAELTRTIVG VVEACHSLGV MHRDLKPENF LFVSKHEDSL LKTIDFGLSM 300
301 FFKPDDVFTD VVGSPYYVAP EVLRKRYGPE ADVWSAGVIV YILLSGVPPF WAETEQGIFE 360
361 QVLHGDLDFS SDPWPSISES AKDLVRKMLV RDPKKRLTAH QVLCHPWVQV DGVAPDKPLD 420
421 SAVLSRMKQF SAMNKFKKMA LRVIAESLSE EEIAGLKEMF NMIDADKSGQ ITFEELKAGL 480
481 KRVGANLKES EILDLMQAAD VDNSGTIDYK EFIAATLHLN KIEREDHLFA AFTYFDKDGS 540
541 GYITPDELQQ ACEEFGVEDV RIEELMRDVD QDNDGRIDYN EFVAMMQKGS ITGGPVKMGL 600
601 EKSFSIALKL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
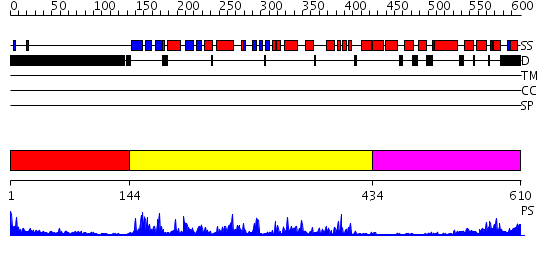
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..143] | 1.83 | No description for 2j63A was found. | |
| 2 | View Details | [144..433] | 76.69897 | CALMODULIN-DEPENDENT PROTEIN KINASE FROM RAT | |
| 3 | View Details | [434..610] | 25.69897 | Regulatory apparatus of Calcium Dependent protein kinase from Arabidopsis thaliana |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)