













| General Information: |
|
| Name(s) found: |
HMA1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 819 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
chloroplast envelope [IDA] |
| Biological Process: |
cellular copper ion homeostasis
[IMP]
calcium ion transport [IGI] zinc ion homeostasis [IMP] response to light intensity [IMP] response to toxin [IGI] |
| Molecular Function: |
ATPase activity
[IMP]
cadmium-transporting ATPase activity [IGI] copper-exporting ATPase activity [ISS] calcium-transporting ATPase activity [IGI] zinc transporting ATPase activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEPATLTRSS SLTRFPYRRG LSTLRLARVN SFSILPPKTL LRQKPLRISA SLNLPPRSIR 60
61 LRAVEDHHHD HHHDDEQDHH NHHHHHHQHG CCSVELKAES KPQKMLFGFA KAIGWVRLAN 120
121 YLREHLHLCC SAAAMFLAAA VCPYLAPEPY IKSLQNAFMI VGFPLVGVSA SLDALMDIAG 180
181 GKVNIHVLMA LAAFASVFMG NALEGGLLLA MFNLAHIAEE FFTSRSMVDV KELKESNPDS 240
241 ALLIEVHNGN VPNISDLSYK SVPVHSVEVG SYVLVGTGEI VPVDCEVYQG SATITIEHLT 300
301 GEVKPLEAKA GDRVPGGARN LDGRMIVKAT KAWNDSTLNK IVQLTEEAHS NKPKLQRWLD 360
361 EFGENYSKVV VVLSLAIAFL GPFLFKWPFL STAACRGSVY RALGLMVAAS PCALAVAPLA 420
421 YATAISSCAR KGILLKGAQV LDALASCHTI AFDKTGTLTT GGLTCKAIEP IYGHQGGTNS 480
481 SVITCCIPNC EKEALAVAAA MEKGTTHPIG RAVVDHSVGK DLPSIFVESF EYFPGRGLTA 540
541 TVNGVKTVAE ESRLRKASLG SIEFITSLFK SEDESKQIKD AVNASSYGKD FVHAALSVDQ 600
601 KVTLIHLEDQ PRPGVSGVIA ELKSWARLRV MMLTGDHDSS AWRVANAVGI TEVYCNLKPE 660
661 DKLNHVKNIA REAGGGLIMV GEGINDAPAL AAATVGIVLA QRASATAIAV ADILLLRDNI 720
721 TGVPFCVAKS RQTTSLVKQN VALALTSIFL AALPSVLGFV PLWLTVLLHE GGTLLVCLNS 780
781 VRGLNDPSWS WKQDIVHLIN KLRSQEPTSS SSNSLSSAH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
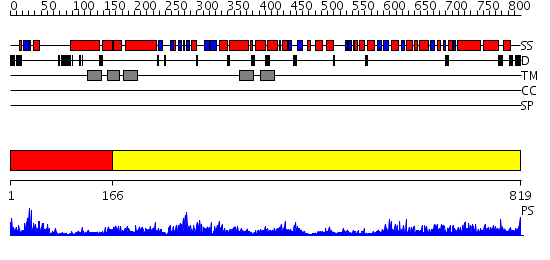
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..165] | 16.0 | Structure characterization of the water soluble region of P-type ATPase CopA from Bacillus subtilis | |
| 2 | View Details | [166..819] | 110.0 | Calcium ATPase, transduction domain A; Calcium ATPase, catalytic domain P; Calcium ATPase; Calcium ATPase, transmembrane domain M |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|