













| General Information: |
|
| Name(s) found: |
ssa2 /
SPCC1739.13
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 647 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
cellular response to heat
[IEP protein folding [ISS protein refolding [ISS |
| Molecular Function: |
ATPase activity
[ISS ATP binding [IEA] unfolded protein binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKSIGIDLG TTYSCVGHFS NNRVEIIAND QGNRTTPSYV AFTDTERLIG DAAKNQVAMN 60
61 PHNTIFDAKR LIGRKFDDPE VQSDMKHWPF KVISKDGKPV LQVEYKGETK TFTPEEISSM 120
121 VLMKMRETAE AYLGGKVTDA VVTVPAYFND SQRQATKDAG LIAGLNVLRI INEPTAAAIA 180
181 YGLDRSNQGE SNVLIFDLGG GTFDVSLLTI EEGIFEVKAT AGDTHLGGED FDSRLVNHFI 240
241 QEFKRKNKKD ITGNARAVRR LRTACERAKR TLSSSAQASI EIDSLFEGID FYTSITRARF 300
301 EELCADLFRK TMEPVERVLR DSKVDKASVN EIVLVGGSTR IPRVQKLVSD FFNGKEPCKS 360
361 INPDEAVAYG AAVQAAVLTG DTSEKTQDLL LLDVAPLSMG IETAGGVMTP LIKRNTTIPT 420
421 KKSEIFSTYS DNQPGVLIQV FEGERARTKD CNLLGKFELS GIPPAPRGVP QIEVTFDVDA 480
481 NGILNVSALE KGTGKTQKIT ITNDKGRLSK EEIDRMVAEA EKYKAEDEAE SGRIQAKNHL 540
541 ESYAYSLRNS LDDPNLKDKV DASDKETVDK AVKETIEWLD SNTTAAKDEF EAKQKELESV 600
601 ANPIMAKIYQ AGGAPGGMPG AAPGAAPGAA PGAAPGGDNG PEVEEVD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
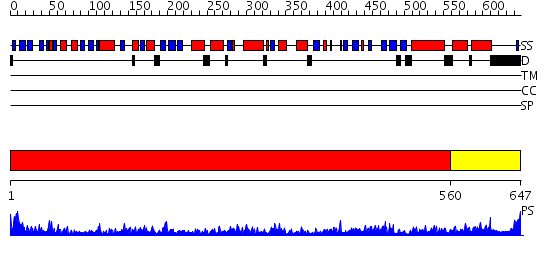
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..559] | 1000.0 | crystal structure of bovine hsc70(aa1-554)E213A/D214A mutant | |
| 2 | View Details | [560..647] | 57.154902 | DnaK |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)