













| General Information: |
|
| Name(s) found: |
nhr-49 /
CE32693
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 501 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
locomotory behavior
[IMP nematode larval development [IMP positive regulation of growth rate [IMP embryonic development ending in birth or egg hatching [IMP regulation of transcription, DNA-dependent [IEA positive regulation of locomotion [IMP |
| Molecular Function: |
DNA binding
[IEA transcription factor activity [IEA zinc ion binding [IEA steroid hormone receptor activity [IEA sequence-specific DNA binding [IEA steroid binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDYFLDASKH IQLNQIDEES SDDFMDLVDP LAEPCAVCGD KSTGTHYGVI SCNGCKGFFR 60
61 RTVLRDQKFT CRFNKRCVID KNFRCACRYC RFQKCVQVGM KREAIQFERD PVGSPTSGAS 120
121 LNGTPFKKDR SPGYENGNSN GVGSNGMGQE NMRTVPQSSS VIDALMEMEA RVNQEMCNRY 180
181 RRSQIFANGS GGSNGNDTDI QQGSDSGASA FAPPNRPCTT EVDLNEISRT TLLLMVEWAK 240
241 TINPFMDLSM EDKIILLKNY APQHLILMPA FRSPDTTRVC LFNNTYMTRD NNTDLNGFAA 300
301 FKTSNITPRV LDEIVWPMRQ LQMREQEFVC LKALAFLHPE AKGLSNSSQI MIRDARNRVL 360
361 KALYAFILDQ MPDDAPTRYG NILLLAPALK ALTQLLIENM TLTKFFGLAE VDSLLSEFIL 420
421 DDINDHSTAP VSLQQHLSSP TTLPTNGVSP LNPAGSVGSV SSVSGITPTG MLSATLAAPL 480
481 AIHPLQSQDS ILNSEQNNHM L |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
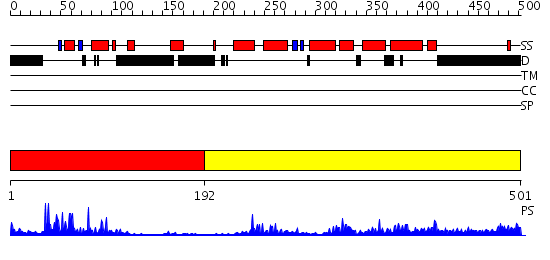
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..191] | 29.221849 | Human Liver Receptor Homologue DNA-Binding Domain (hLRH-1 DBD) in Complex with dsDNA from the hCYP7A1 Promoter | |
| 2 | View Details | [192..501] | 51.09691 | Crystal Structure of the RARbeta/RXRalpha Ligand Binding Domain Heterodimer in Complex with 9-cis Retinoic Acid and a Fragment of the TRAP220 Coactivator |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)