













| General Information: |
|
| Name(s) found: |
atgp-1 /
CE32183
[WormBase]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 613 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA plasma membrane [IDA |
| Biological Process: |
carbohydrate metabolic process
[IEA trehalose catabolic process [IEA |
| Molecular Function: |
alpha-amylase activity
[IEA catalytic activity [IEA calcium ion binding [IEA cation binding [IEA alpha,alpha-phosphotrehalase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATKDALIAE EGTNFGGSSS GDIAVFDKTA EVVNFELEPK PIGLTKEQLE KYRNDPFWKP 60
61 VRTILFVLFW LAWVLMFAGA IAIVVLSPKC AEKQKPDWWQ TKVSYQLLTA TFYDSDNDGV 120
121 GDFAGISQKI DFLRKIGVTT VYPTPVIKIH KDEYFNSYDV VDHNSVDERF GTEEQFKELI 180
181 DTVHNRAMYL VMDLPVSTID LSHPWFESRD ESKFVIAKPT DPGFNESNFY PFHGANNIKY 240
241 LGYPSSQNPV LNWKNADVKS TINGAIQKFL DLGVDGFHID HISQLAVDSK GRPNHDEAVK 300
301 VLEELTKSVQ IYVESRDELV DKKIVLFSSL KDVEDLHVKA TETGLLHYVI DNSFANLDEK 360
361 KCQPHVAKCV HDALNAAYQR HEVAKYTPHW QFSNSEASRL ASRFESATAH LLSFLQLTLP 420
421 GANSVYYGQE YGLKDAMSKN GEFKQMGVMQ WYPAGKDHHG FSGNDAPIFF PESDDKLELD 480
481 NYNTQFETEN SALKIYRKLA KLRQRDEALI VGETVRDELI NEDVILFSRY VQAVNNTATG 540
541 STFIVALNFG DKEQKIDFSI EPASKLIPSN KDLLNAEISV VTANVTDYRV RDHHNFVESP 600
601 LVLPPKQAVL LKL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
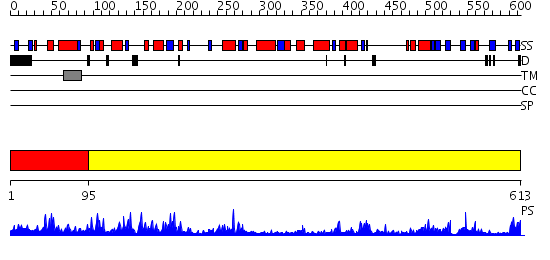
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..94] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [95..613] | 111.0 | CRYSTAL STRUCTURE OF B. CEREUS OLIGO-1,6-GLUCOSIDASE |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|