













| General Information: |
|
| Name(s) found: |
mtm-6 /
CE32050
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 782 amino acids |
Gene Ontology: |
|
| Cellular Component: |
apical plasma membrane
[IDA]
|
| Biological Process: |
phospholipid dephosphorylation
[IEA protein amino acid dephosphorylation [IEA endocytosis [IMP dephosphorylation [IEA positive regulation of multicellular organism growth [IMP |
| Molecular Function: |
zinc ion binding
[IEA inositol or phosphatidylinositol phosphatase activity [IEA protein tyrosine phosphatase activity [IEA phosphatase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSTPSSVTT PLSAWWNNMS LSGIYPSIST STPSSSSSWS SHFVFGGHHH HTNNNTANTP 60
61 QSTVPPSALS SGYTEDITVA GSLLKDKING LVDSLMGRES VHSGCGYGED GRIAMMVDKV 120
121 CLVDRLGCQE NLVGTVHVTT THIIFRAENG SKELWLATGL ISSVEKGTLT AAGCMLVIRC 180
181 KHFQVITLLI SRDKSCQDLY ETLQRAAKPV SVNVTELLAF ENREPVEDVR GWRRLDWNSE 240
241 MTRQGITKSQ WTESNINEGY TICDTYPNKL WFPTAASTSV LLGSCKFRSR GRLPVLTYFH 300
301 QQTEAALCRC AQPLTGFSAR CVEDEKLMEL VGKANTNSDN LFLVDTRPRV NAMVNKVQGK 360
361 GFEDERNYSN MRFHFFDIEN IHVMRASQAR LLDAVTKCRD VTEYWKTLEA SGWLKHVRSV 420
421 VECSLFLAES ISRGTSCVVH CSDGWDRTSQ VVALCQLLLD PYYRTIHGFQ VLIEKDWLGF 480
481 GHKFDDRCGH VGALNDEAGK EVSPIFTQWL DCIWQIMQQK PRAFQFNERY LIEMHEHVYS 540
541 CQFGTFIGNC DKDRRDLNLS KRTKSLWTWM DARHDDYMNP FYSPTAHVAL LDLDTRAARF 600
601 TVWTAMYNRF DNGLQPRERL EDLTMAAMEH VGVLESHVAQ LRTRLAELKT QQNQQITSTN 660
661 TPTNMVDSGM SSATDDLKNL SLSHPLDPLS STLPILERAT SQESGVMDSS LYYPDEALTK 720
721 YSLKWQPLRG ADRCSNPACR GEFSSTIERR IHCHLCGMIF CRRCLKVSAD ERERVCDKCK 780
781 TD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
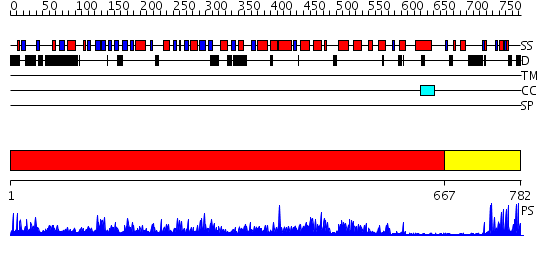
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..666] | 1000.0 | Crystal Structure of Myotubularin-related protein 2 complexed with phosphate | |
| 2 | View Details | [667..782] | 15.045757 | Hrs |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.54 |
Source: Reynolds et al. (2008)