













| General Information: |
|
| Name(s) found: |
pif-1 /
CE30311
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 677 amino acids |
Gene Ontology: |
|
| Cellular Component: |
exodeoxyribonuclease V complex
[IEA |
| Biological Process: | NONE FOUND |
| Molecular Function: |
nucleotide binding
[IEA nucleoside-triphosphatase activity [IEA exodeoxyribonuclease V activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKSLQTDEN GGVSESPSNC TYCYTLECSL RIESTSSIKK KTPISSKSAI MTVGRNAQRK 60
61 IHLQIELKTT ATGQPAVVCY DVTDAVVHLQ SVANGKCTVE IPSLSLMFQM FNCAPRKLNV 120
121 FMKSLQAKLD IMKMEKSPIS AVPRQFSRPP AVFSVLSPLT ISEMRKVKKL REPSALARPS 180
181 KEATTPKRRT SSMNLLAGGL ENRIMNRSIG LKRTTSFARD DREKAETLVS LKSFKDAPAI 240
241 SERIQLSDEQ KSVVRCVINS RTSVFFTGSA GTGKSVILRR IIEMLPAGNT YITAATGVAA 300
301 SQIGGITLHA FCGFRYENST PEQCLKQVLR QNHMVRQWKQ CSHLIIDEIS MIDRDFFEAL 360
361 EYVARTVRNN DKPFGGIQLI ITGDFFQLPP VSKDEPVFCF ESEAWSRCIQ KTIVLKNVKR 420
421 QNDNVFVKIL NNVRVGKCDF KSADILKESS KNQFPSSVIP TKLCTHSDDA DRINSSSIET 480
481 TQGDAKTFHA YDDESFDTHA KARTLAQKKL VLKVGAQVML IKNIDVIKGL CNGSRGFVEK 540
541 FSENGNPMIR FVSQADASIE IRRSKFSVRI PGSDAPLIRR QLPLQLAWAI SIHKSQGMTL 600
601 DCAEISLERV FADGQAYVAL SRARSLAAIR IIGFDASCVR ANSKVIDFYK SIEAECDDEQ 660
661 DWEAPAAGPR LKRVRSI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
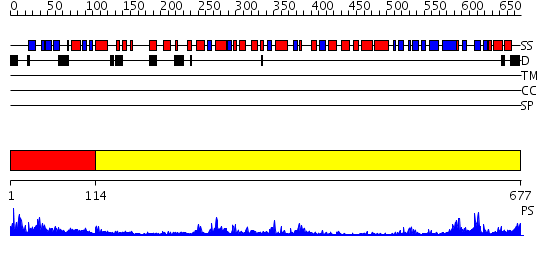
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..113] | 3.522879 | Crystal Structure Analysis of ClpB | |
| 2 | View Details | [114..677] | 46.0 | RecBCD:DNA complex |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)