













| General Information: |
|
| Name(s) found: |
pink-1 /
CE28904
[WormBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 641 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA]
mitochondrion [IDA] |
| Biological Process: |
protein amino acid phosphorylation
[IEA |
| Molecular Function: |
ATP binding
[IEA protein tyrosine kinase activity [IEA protein kinase activity [IEA protein serine/threonine kinase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSMKRFGKAA YRIANELVAK GGRLPIFQRF LPRIFPATYN LGVHVVLKKA PFPRQNALRI 60
61 ARLVTRHGRV FRPFSSVIIE RHRFQNQNDW RRKFQPIRKE LPRNVDLVER IRQIFGNSLR 120
121 YNEDLKSTEW PNRIDSYEFG EFLGQGCNAA VYSARLANSD AESSGNTHYG AGFNEVTNIL 180
181 AEIPPVSKVA QKKFPLAIKL MFNFEHDRDG DAHLLKSMGN ELAPYPNAAK LLNGQMGTFR 240
241 PLPAKHPNVV RIQTAFIDSL KVLPDAIERY PDALHTARWY ESIASEPKTM YVVMRRYRQT 300
301 LHEYVWTRHR NYWTGRVIIA QLLEACTYLH KHKVAQRDMK SDNILLEYDF DDEIPQLVVA 360
361 DFGCALACDN WQVDYESDEV SLGGNAKTKA PEIATAVPGK NVKVNFEMAD TWAAGGLSYE 420
421 VLTRSNPFYK LLDTATYQES ELPALPSRVN FVARDVIFDL LKRDPNERVK PNIAANALNL 480
481 SLFRMGEDVK QMMEKCGISQ MTTLLAGSSK VLSQKINSRL DKVMNLITAE TIMANLAPHL 540
541 ISRAERQLRA TFLSRMNRED IWRSLQYFFP AGVQLDTPAT SSDCLETISS LMSSFSNDSE 600
601 NYEKQQKPAK NGYNNVPLLL RNVIRTDADG INGIVHRVRS K |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
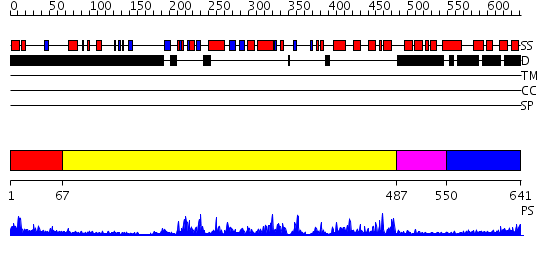
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..66] | 12.045757 | No description for 2uvmA was found. | |
| 2 | View Details | [67..486] | 56.0 | CHICKEN SRC TYROSINE KINASE | |
| 3 | View Details | [487..549] | 73.30103 | Crystal Structure of ROCK 1 bound to fasudil | |
| 4 | View Details | [550..641] | 6.39794 | No description for 2ux0A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)