













| General Information: |
|
| Name(s) found: |
CE27852
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 351 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA |
| Biological Process: |
lysine biosynthetic process
[IEA arginine biosynthetic process [IEA lysine biosynthetic process via diaminopimelate [IEA proteolysis [IEA cellular amino acid metabolic process [IEA |
| Molecular Function: |
hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides
[IEA zinc ion binding [IEA metal ion binding [IEA dipeptidase activity [IEA protein dimerization activity [IEA succinyl-diaminopimelate desuccinylase activity [IEA aminoacylase activity [IEA acetylornithine deacetylase activity [IEA metallopeptidase activity [IEA cobalt ion binding [IEA hydrolase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTIKWEVEVE KRLLEYMSHS STTGNEAAFA DVVAKDLEEN GWTVYKQSIP NSDRYNIYAT 60
61 FRNSDPKHVK VLLNTHLDTV PPYFPPTQDE QNIYGNGSND AKGQLAAMVT AATIISKTDE 120
121 DVARALGLLF VVGEEFDHIG MIEANKLEIL PEYLLVGEPT ELKFGTIQKG ALKVKLTVTG 180
181 QAGHSGYPNS GSSAIHKMIE VLHDVQLAKW PCDATNGATT YNIGKISGGQ ALNAWAANCE 240
241 ADIFFRVVTS VKDIQNQLLA LVNGRAEVSL LSFNDPVILD VPPIEAELDH VSFNTDIAYF 300
301 DARDKVKAKY LFGGGSIKNA HSKNEFIPKD ELHKCTATLV KLVNHLYLEH Y |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
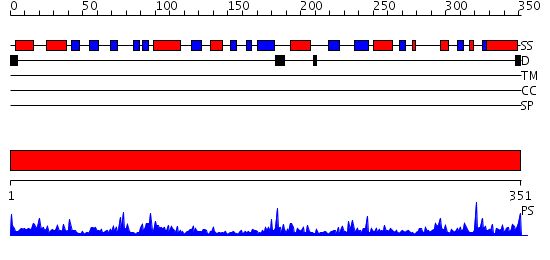
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..351] | 65.69897 | Crystal structure of succinyl diaminopimelate desuccinylase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)