













| General Information: |
|
| Name(s) found: |
hsp-60 /
CE27244
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 568 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA |
| Biological Process: |
embryonic development ending in birth or egg hatching
[IMP cellular protein metabolic process [IEA protein folding [IEA reproduction [IMP |
| Molecular Function: |
ATP binding
[IEA protein binding [IEA unfolded protein binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLRLARKGLQ TAVVRSYAKD VKFGAEGRQA MLVGVNLLAD AVSVTMGPKG RNVIIEQSWG 60
61 SPKITKDGVT VAKSIDLKDK YQNLGAKLIQ DVANKANEEA GDGTTCATVL ARAIAKEGFE 120
121 SIRQGGNAVE IRRGVMNAVE VVVAELKKIS KKVTTPEEIA QVATISANGD TVVGNLISDA 180
181 MKKVGTTGVI TVKDGKTLND ELELIEGMKF DRGYISPYFI TSAKGAKVEY EKALVLLSEK 240
241 KISQVQDIVP ALELANKLRR PLVIIAEDVD GEALTTLVLN RLKVGLQVVA IKAPGFGDNR 300
301 KNTLKDMGIA TGATIFGDDS NLIKIEDITA NDLGEVDEVT ITKDDTLLLR GRGDQTEIEK 360
361 RIEHITDEIE QSTSDYEKEK LNERLAKLSK GVAVLKIGGG SEVEVGEKKD RVTDALCATR 420
421 AAVEEGIVPG GGVALLRSLT ALKNYKAANE DQQIGVNIVK KALTQPIATI VKNAGLEPSS 480
481 IIDEVTGNSN TSYGYDALNG KFVDMFEAGI IDPTKVVRTA LQDASGVASL LATTECVVTE 540
541 IPKEEAVGGP AGGMGGMGGM GGMGGMGF |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | hcp1 IP from december2004 | Cheeseman, I.M., et al. (2005) | |
| View Run | hcp2 ip from december | Cheeseman, I.M., et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
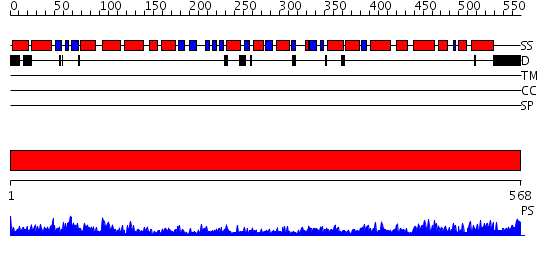
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..568] | 167.0 | GroEL, E domain; GroEL, A domain; GroEL, I domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.78 |
Source: Reynolds et al. (2008)