













| General Information: |
|
| Name(s) found: |
vab-3 /
CE24899
[WormBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 455 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
multicellular organismal development
[IEA cell fate commitment [IMP anterior/posterior pattern formation [IMP regulation of transcription, DNA-dependent [IEA tail tip morphogenesis [IMP regulation of cell adhesion [IMP positive regulation of transcription from RNA polymerase II promoter [IMP |
| Molecular Function: |
transcription factor activity
[IEA sequence-specific DNA binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDAGHTGVN QLGGVFVNGR PLPDATRQRI VDLAHKGCRP CDISRLLQVS NGCVSKILCR 60
61 YYESGTIRPR AIGGSKPRVA TSDVVEKIED YKRDQPSIFA WEIRDKLLAD NICNNETIPS 120
121 VSSINRVLRN LAAKKEQVTM QTELYDRIRI VDNFPYNSSW YGQWPIPMNG AVGLNPFVPA 180
181 PLIEPKTEGE FEKDEDQKPP TEPEDDAAAR MRLKRKLQRN RTSFTQVQIE SLEKEFERTH 240
241 YPDVFARERL AQKIQLPEAR IQVWFSNRRA KWRREEKMRN KRSSGTMDSS LSNGTPTPTP 300
301 GSVTGSNMTN PIGSPASTPN RFPSNNSANL PTTNFVPQTS QMYAGLSQPA MDPYSFGIAN 360
361 GFSMAPYPQV TDFQPHHMFQ GRSPYDFPYP RMPTNGHGFQ QSMSPATTAV GDIPTLSSGM 420
421 SLPVSAVLNS IDPSLTHSQM HELSDLTQEH YWRPQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
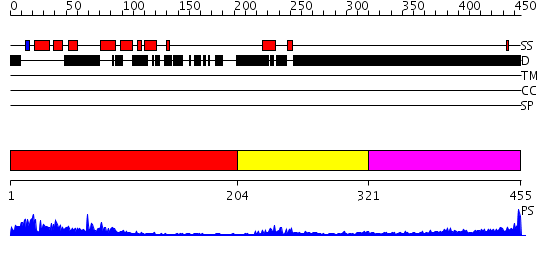
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..203] | 50.30103 | Pax-5 | |
| 2 | View Details | [204..320] | 12.522879 | Solution structure of the homeobox domain of the human paired box protein Pax-6 | |
| 3 | View Details | [321..455] | 4.374996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)