













| General Information: |
|
| Name(s) found: |
acs-1 /
CE20812
[WormBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 623 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
propionate catabolic process, 2-methylcitrate cycle
[IEA teichoic acid biosynthetic process [IEA embryonic development ending in birth or egg hatching [IMP metabolic process [IEA menaquinone biosynthetic process [IEA siderophore biosynthetic process [IEA reproduction [IMP |
| Molecular Function: |
D-alanine-poly(phosphoribitol) ligase activity
[IEA AMP binding [IEA acetate-CoA ligase activity [IEA catalytic activity [IEA propionate-CoA ligase activity [IEA o-succinylbenzoate-CoA ligase activity [IEA (2,3-dihydroxybenzoyl)adenylate synthase activity [IEA ligase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQVAAMDLR VLTQFDIASL EEDRKKMLYE EPISLEEAAL NANDVMVAPS RKSYVHGCST 60
61 VPLLFETVGD RLRSAVDQVP DKEFLIFKRE GIRKTYSQVA TDAENLACGL LHLGLKKGDR 120
121 IGIWGPNTYE WTTTQFASAL AGMVLVNINP SYQSEELRYA IEKVGIRALI TPPGFKKSNY 180
181 YQSIKDILPE VTLKEPGKSG ITSRNFTCFQ HLIMFDEEDK IYPGAWKYTD VMKMGTEEDR 240
241 HHLSKIERET QPDDSLNIQY TSGTTGQPKG ATLTHHNVLN NAFFVGLRAG YSEKKTIICI 300
301 PNPLYHCFGC VMGVLAALTH LQTCVFPAPS FDALAALQAI HEEKCTALYG TPTMFIDMIN 360
361 HPEYANYNYD SIRSGFIAGA PCPITLCRRL VQDMHMTDMQ VCYGTTETSP VSFMSTRDDP 420
421 PEQRIKSVGH IMDHLEAAIV DKRNCIVPRG VKGEVIVRGY SVMRCYWNSE EQTKKEITQD 480
481 RWYHTGDIAV MHDNGTISIV GRSKDMIVRG GENIYPTEVE QFLFKHQSVE DVHIVGVPDE 540
541 RFGEVVCAWV RLHESAEGKT TEEDIKAWCK GKIAHFKIPR YILFKKEYEF PLTVTGKVKK 600
601 FEIREMSKIE LGLQQVVSHF SEL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
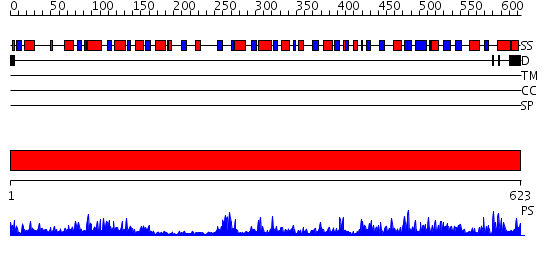
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..623] | 158.0 | No description for 2p2bA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)