













| General Information: |
|
| Name(s) found: |
mtm-6 /
CE16100
[WormBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 676 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
phospholipid dephosphorylation
[IEA protein amino acid dephosphorylation [IEA endocytosis [IMP dephosphorylation [IEA positive regulation of multicellular organism growth [IMP |
| Molecular Function: |
zinc ion binding
[IEA inositol or phosphatidylinositol phosphatase activity [IEA protein tyrosine phosphatase activity [IEA phosphatase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRFEDIGISK VDKVCLVDRL GCQENLVGTV HVTTTHIIFR AENGSKELWL ATGLISSVEK 60
61 GTLTAAGCML VIRCKHFQVI TLLISRDKSC QDLYETLQRA AKPVSVNVTE LLAFENREPV 120
121 EDVRGWRRLD WNSEMTRQGI TKSQWTESNI NEGYTICDTY PNKLWFPTAA STSVLLGSCK 180
181 FRSRGRLPVL TYFHQQTEAA LCRCAQPLTG FSARCVEDEK LMELVGKANT NSDNLFLVDT 240
241 RPRVNAMVNK VQGKGFEDER NYSNMRFHFF DIENIHVMRA SQARLLDAVT KCRDVTEYWK 300
301 TLEASGWLKH VRSVVECSLF LAESISRGTS CVVHCSDGWD RTSQVVALCQ LLLDPYYRTI 360
361 HGFQVLIEKD WLGFGHKFDD RCGHVGALND EAGKEVSPIF TQWLDCIWQI MQQKPRAFQF 420
421 NERYLIEMHE HVYSCQFGTF IGNCDKDRRD LNLSKRTKSL WTWMDARHDD YMNPFYSPTA 480
481 HVALLDLDTR AARFTVWTAM YNRFDNGLQP RERLEDLTMA AMEHVGVLES HVAQLRTRLA 540
541 ELKTQQNQQI TSTNTPTNMV DSGMSSATDD LKNLSLSHPL DPLSSTLPIL ERATSQESGV 600
601 MDSSLYYPDE ALTKYSLKWQ PLRGADRCSN PACRGEFSST IERRIHCHLC GMIFCRRCLK 660
661 VSADERERVC DKCKTD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
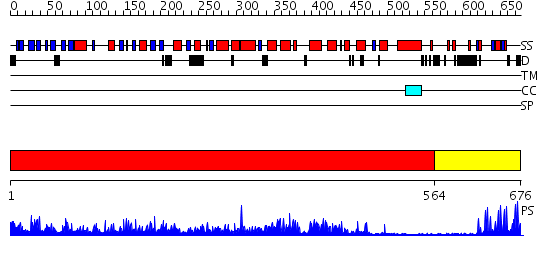
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..563] | 174.0 | Crystal Structure of Myotubularin-related protein 2 complexed with phosphate | |
| 2 | View Details | [564..676] | 12.0 | Hrs |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)