













| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNSNGPLMEH GKRRVAYYYD SNIGNYYYGQ GHVMKPHRIR MTHHLVLNYG LYRNLEIFRP 60
61 FPASFEDMTR FHSDEYMTFL KSANPDNLKS FNKQMLKFNV GEDCPLFDGL YEFCQLSSGG 120
121 SLAAATKLNK QKVDIAINWM GGLHHAKKSE ASGFCYTNDI VLGILELLKY HKRVLYVDID 180
181 VHHGDGVEEA FYTTDRVMTV SFHKYGDFFP GTGDLKDIGA GKGKLYSVNV PLRDGITDVS 240
241 YQSIFKPIMT KVMERFDPCA VVLQCGADSL NGDRLGPFNL TLKGHGECAR FFRSYNVPLM 300
301 MVGGGGYTPR NVARCWTYET SIAVDKEVPN ELPYNDYFEY FGPNYRLHIE SSNAANENSS 360
361 DMLAKLQTDV IANLEQLTFV PSVQMRPIPE DALSALNDDS LIADQANPDK RLPPQITDGM 420
421 IQDDGDFYDG EREGDDRRNE SDAKRAAQFE SEGGEKRQKT E |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | hcp1 IP from december2004 | Cheeseman, I.M., et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
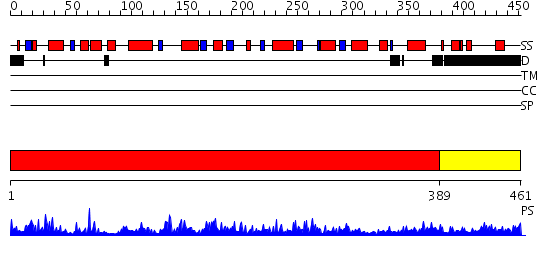
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..388] | 94.0 | Crystal Structure of human HDAC8 complexed with Trichostatin A | |
| 2 | View Details | [389..461] | 1.097991 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)