













| General Information: |
|
| Name(s) found: |
hsp-4 /
CE07244
[WormBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 657 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum
[IEA |
| Biological Process: |
embryonic development
[IMP endoplasmic reticulum unfolded protein response [IEP nematode larval development [IMP cell morphogenesis [IEA embryonic development ending in birth or egg hatching [IMP positive regulation of growth rate [IMP protein folding [IEA response to stress [IDA growth [IMP reproduction [IMP iron-sulfur cluster assembly [IEA |
| Molecular Function: |
ATPase activity
[IEA ATP binding [IEA unfolded protein binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKVFSLILIA FVANAYCDEG ASTEKEEKMG TIIGIDLGTT YSCVGVFKNG RVEIIANDQG 60
61 NRITPSYVAF SGDQGERLIG DAAKNQLTIN PENTIFDAKR LIGRFYNEKT VQQDIKHWPF 120
121 KIVDKSNKPN VEVKVGSETK QFTPEEVSAM VLTKMKQIAE SYLGHEVKNA VVTVPAYFND 180
181 AQKQATKDAG SIAGLNVVRI INEPTAAAIA YGLDKKDGER NILVFDLGGG TFDVSLLTID 240
241 SGVFEVLATN GDTHLGGEDF DQRVMEYFIK LYKKKSGKDL RKDNRAVQKL RREVEKAKRA 300
301 LSTQHQTKIE IESLFDGEDF SETLTRAKFE ELNMDLFRAT LKPVQKVLED ADMKKTDVHE 360
361 IVLVGGSTRI PKVQQLIKDY FNGKEPSRGI NPDEAVAYGA AVQAGVIGGV ENTGDVVLLD 420
421 VNPLTLGIET VGGVMTKLIG RNTVIPTKKS QVFSTAADSQ SAVSIVIYEG ERPMVMDNHK 480
481 LGNFDVTGIP PAPRGVPQIE VTFEIDVNGI LHVSAEDKGT GNKNKLTITN DHNRLSPEDI 540
541 ERMINDADKF AADDQAQKEK VESRNELEAY AYQMKTQIAD KEKLGGKLTD EDKVSIESAV 600
601 ERAIEWLGSN QDASTEENKE QKKELESVVQ PIVSKLYSAG GQGEQASEEP SEDHDEL |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) | |
| View Run | hcp1 IP from december2004 | Cheeseman, I.M., et al. (2005) | |
| View Run | hcp2 ip from december | Cheeseman, I.M., et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
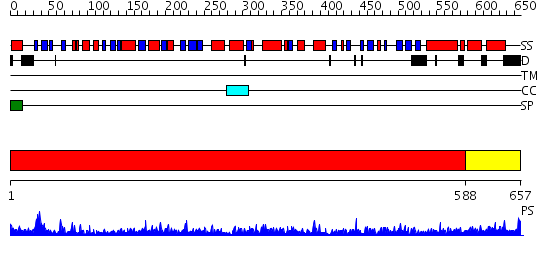
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..587] | 1000.0 | crystal structure of bovine hsc70(aa1-554)E213A/D214A mutant | |
| 2 | View Details | [588..657] | 54.221849 | DnaK |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|