













| General Information: |
|
| Name(s) found: |
tag-38 /
CE06695
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 542 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
glutamate metabolic process
[IEA metabolic process [IEA proteolysis [IEA carboxylic acid metabolic process [IEA |
| Molecular Function: |
pyridoxal phosphate binding
[IEA zinc ion binding [IEA transaminase activity [IEA glutamate decarboxylase activity [IEA carboxy-lyase activity [IEA metallopeptidase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDFALEQYHS AKDLLIFELR KFNPIVLVSS TIVATYVLTN LRHMHLDEMG IRKRLSTWFF 60
61 TTVKRVPFIR KMIDKQLNEV KDELEKSLRI VDRSTEYFTT IPSHSVGRTE VLRLAAIYDD 120
121 LEGPAFLEGR VSGAVFNRED DKDEREMYEE VFGKFAWTNP LWPKLFPGVR IMEAEVVRMC 180
181 CNMMNGDSET CGTMSTGGSI SILLACLAHR NRLLKRGEKY TEMIVPSSVH AAFFKAAECF 240
241 RIKVRKIPVD PVTFKVDLVK MKAAINKRTC MLVGSAPNFP FGTVDDIEAI GQLGLEYDIP 300
301 VHVDACLGGF LLPFLEEDEI RYDFRVPGVS SISADSHKYG LAPKGSSVVL YRNKELLHNQ 360
361 YFCDADWQGG IYASATMEGS RAGHNIALCW AAMLYHAQEG YKANARKIVD TTRKIRNGLS 420
421 NIKGIKLQGP SDVCIVSWTT NDGVELYRFH NFMKEKHWQL NGLQFPAGVH IMVTMNHTHP 480
481 GLAEAFVADC RAAVEFVKSH KPSESDKTSE AAIYGLAQSI PDRSLVHEFA HSYIDAVYAL 540
541 TE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
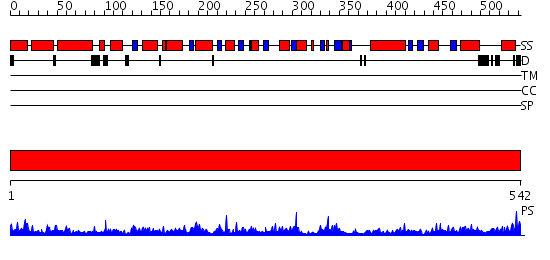
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..542] | 50.30103 | Crystal structure of the complex of Escherichia coli GADA with glutarate at 2.05 A resolution |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.72 |
Source: Reynolds et al. (2008)