













| General Information: |
|
| Name(s) found: |
CE06652
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 536 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
positive regulation of growth rate
[IMP cellular amino acid metabolic process [IEA |
| Molecular Function: |
oxidoreductase activity
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLSTLSRGAR SVAVRSYSAA TLDAHSQVLD EQKPMEEQVN PSFYKMVDFY FNKGAEVIAP 60
61 KLAEELKSNS LSQKDKKNLV SGILGAIKPV NKVLYITFPI RRDNGEFEVI EAWRAQHSEH 120
121 RTPTKGGIRY SLDVCEDEVK ALSALMTYKC AVVDVPFGGA KGGVKIDPKQ YTDYEIEKIT 180
181 RRIAIEFAKK GFLGPGVDVP APDMGTGERE MGWIADTYAQ TIGHLDRDAS ACITGKPIVS 240
241 GGIHGRVSAT GRGVWKGLEV FTNDADYMKM VGLDTGLAGK TAIIQGFGNV GLHTHRYLHR 300
301 AGSKVIGIQE YDCAVYNPDG IHPKELEDWK DANGTIKNFP GAKNFDPFTE LMYEKCDIFV 360
361 PAACEKSIHK ENASRIQAKI IAEAANGPTT PAADRILLAR GDCLIIPDMY VNSGGVTVSY 420
421 FEWLKNLNHV SYGRLTFKYD EEANKMLLAS VQESLSKAVG KDCPVEPNAA FAAKIAGASE 480
481 KDIVHSGLEY TMQRSGEAII RTAHKYNLGL DIRTAAYANS IEKVYNTYRT AGFTFT |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | hcp1 IP from december2004 | Cheeseman, I.M., et al. (2005) | |
| View Run | hcp2 ip from december | Cheeseman, I.M., et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
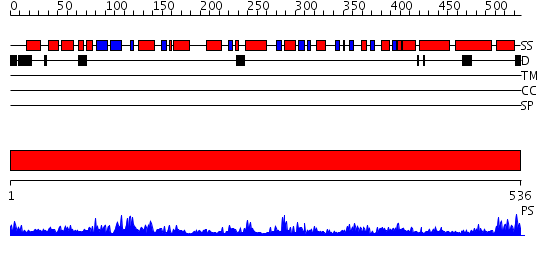
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..536] | 141.0 | Glutamate dehydrogenase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.66 |
Source: Reynolds et al. (2008)