













| General Information: |
|
| Name(s) found: |
pcca-1 /
CE04451
[WormBase]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 724 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA acetyl-CoA carboxylase complex [IEA |
| Biological Process: |
sodium ion transport
[IEA gluconeogenesis [IEA metabolic process [IEA peptidoglycan biosynthetic process [IEA protein modification process [IEA purine base biosynthetic process [IEA fatty acid biosynthetic process [IEA |
| Molecular Function: |
oxaloacetate decarboxylase activity
[IEA ATP binding [IEA D-alanine-D-alanine ligase activity [IEA acetyl-CoA carboxylase activity [IEA pyruvate carboxylase activity [IEA biotin binding [IEA ligase activity [IEA hydroxymethyl-, formyl- and related transferase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLRAASSIRA VANRGLATAA VARPGVPLDE REGKEIYTTV GIDYNEPKFD KILIANRGEI 60
61 ACRVIKTARA MGIKTVAVHS DVDSNSLHVK MADEAVCVGE APTAKSYLRA DRILQAVEDT 120
121 GAQAVHPGYG FLSENTKFAA ELEKAGAKFI GPNSKAILDM GDKIHSKKIA TAARVSMIPG 180
181 YDGEIADEDM CVKVSRDIGY PVMIKASAGG GGKGMRVAWN DKQAREGYRL SKQEAASSFG 240
241 DDRMLVEKFI DNPRHIEMQV LCDKHGNALW LNERECSIQR RNQKVIEEAP SSFVPPEMRR 300
301 KMGEQAVQLA KAVGYDSAGT VEFLVDSQRN FYFLEMNTRL QVEHPITECI TGIDIVQQML 360
361 RVSYGHPLPI TQEQVPLNGW AFESRVYAED PYKGFGLPSV GRLSRYVEPK HVDGVRCDSG 420
421 IREGSEISIY YDPLICKLVT HGDNREQALN RMQEALDNYV IRGVTHNIPL LRDIVQEKRF 480
481 RTGDITTKYL PEVYPEGFQG TSLSPKEQDV VIAFASALNA RKLARANQFL NQNKQRSTHV 540
541 ASFSKTYKFV SSLPVKEGER PTEHAVEVEF VEGSANKAQV RIGGKTVTIS GDLNLSHPVN 600
601 SIEVDGEHIT TQIVGKRAGE ITVLYKGTPF KVKVLPEQAV KYLQYMKEKA KVDLSTVVLS 660
661 PMPGAIKNVN VKPGDMVSEG QELVVMEAMK MQNSLHAGKT GRVKAVNVKV GATVDEGEVL 720
721 VELE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
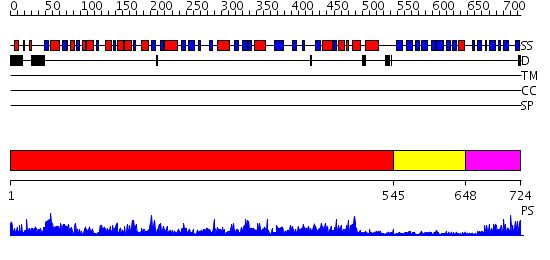
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..544] | 139.0 | No description for 2gpwA was found. | |
| 2 | View Details | [545..647] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [648..724] | 15.09691 | No description for 2d5dA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|