













| General Information: |
|
| Name(s) found: |
lin-39 /
CE03975
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 253 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
negative regulation of vulval development
[IMP regulation of cell division [IMP positive regulation of vulval development [IGI vulval development [IEP regulation of cell fate specification [IMP locomotory behavior [IMP anterior/posterior pattern formation [IMP regulation of transcription, DNA-dependent [IEA oviposition [IMP Ras protein signal transduction [IEP plasma membrane fusion [IMP cell migration [IMP |
| Molecular Function: |
DNA binding
[IEA transcription factor activity [IEA sequence-specific DNA binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTSTSPSST DAPRATAPES SSSSSSSSSS SSSTSSVGAS GIPSSSELSS TIGYDPMTAS 60
61 AALSAHFGSY YDPTSSSQIA SYFASSQGLG GPQYPILGDQ SLCYNPSVTS THHDWKHLEG 120
121 DDDDDKDDDK KGISGDDDDM DKNSGGAVYP WMTRVHSTTG GSRGEKRQRT AYTRNQVLEL 180
181 EKEFHTHKYL TRKRRIEVAH SLMLTERQVK IWFQNRRMKH KKENKDKPMT PPMMPFGANL 240
241 PFGPFRFPLF NQF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
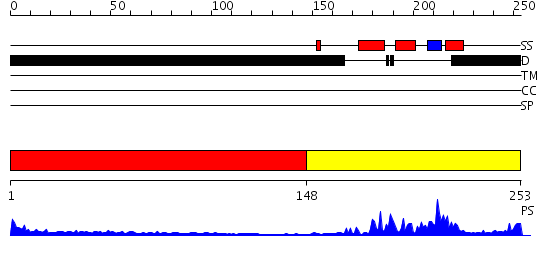
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..147] | 16.122996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [148..253] | 28.69897 | DETERMINATION OF THE THREE-DIMENSIONAL STRUCTURE OF THE ANTENNAPEDIA HOMEODOMAIN FROM DROSOPHILA IN SOLUTION BY 1H NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.66 |
Source: Reynolds et al. (2008)