













| General Information: |
|
| Name(s) found: |
nas-10 /
CE03479
[WormBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 560 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
proteolysis
[IEA |
| Molecular Function: |
astacin activity
[IEA metallopeptidase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYTITDLKTI NRKQKQAYLQ EKLICSRHCW LTHTTSKHIN IQWTTLIPHI RCRGFNNGPH 60
61 PNSRPGSRPN SPLGDIFGNI NGMVKGITDQ IGKIAQGLDV NNDLGKMAHG PPPPQSEWVE 120
121 HARRFCRRFP GHPKCRGQLP QFNDIGSMLN GILVDSGKWL PKVPFINIRD PLSGINSDLK 180
181 NALNGIQVQF GQISQQFANN IRNICQQMNC KQQQQKNVQM KQAILKQTVD FEKKVFGNNV 240
241 ADKMNLRFDR TLQLKQALLE KAQLKGVVAP EDNGVFDKDL LLTETQANFM LNELGKGGEG 300
301 AIPMPGSAKA KRASIFFEQN LIQKWPSTSP IPYTFDSSLD NLDQNDVRGA ISEIEQKTCI 360
361 RFKYFASPPK GNHINYQKVN SPSFCGLSYI GRVEPANPVY LSFQCGNGRG IAVHETMHAL 420
421 GVNHQHLRMD RDKHIKVDWS NINPQQYDAF VVADSKLYTT YGVKYAYDSI MHYNAYTGAV 480
481 NIAKPTMIPL VNQQANIGLL GQRAKMSNAD VEILNKMYCK SAGCDDKNVY CGAWALQDLC 540
541 NNPNHNVWMR SNCRKSCNFC |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
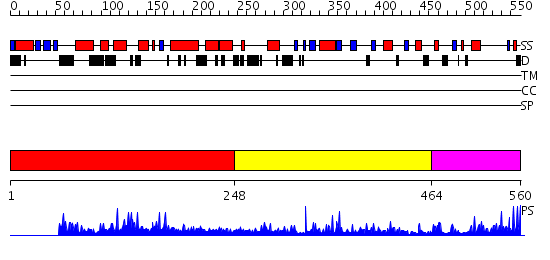
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..247] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [248..463] | 33.09691 | Gelatinase A (MMP-2); Gelatinase A (MMP-2), C-terminal domain; Gelatinase A; Gelatinase A (MMP-2) type II modules | |
| 3 | View Details | [464..560] | 41.69897 | STRUCTURE OF ASTACIN AND IMPLICATIONS FOR ACTIVATION OF ASTACINS AND ZINC-LIGATION OF COLLAGENASES |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)