













| General Information: |
|
| Name(s) found: |
drs-1 /
CE00015
[WormBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 531 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA |
| Biological Process: |
tRNA aminoacylation for protein translation
[IEA nematode larval development [IMP embryonic development ending in birth or egg hatching [IMP asparaginyl-tRNA aminoacylation [IEA lysyl-tRNA aminoacylation [IEA aspartyl-tRNA aminoacylation [IEA reproduction [IMP growth [IMP translation [IEA |
| Molecular Function: |
nucleotide binding
[IEA nucleic acid binding [IEA ATP binding [IEA aminoacyl-tRNA ligase activity [IEA lysine-tRNA ligase activity [IEA asparagine-tRNA ligase activity [IEA aspartate-tRNA ligase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MADAAEGEQP KLSKKELNKL ARKAKKDEKA GEKGGNQQQA AAMDQEDASK DFYGSYGLVN 60
61 SKEKKVLNFL KVKEINVSNA TKDVWVRGRI HTTRSKGKNC FLVLRQGVYT VQVAMFMNEK 120
121 ISKQMLKFVS SISKESIVDV YATINKVDNP IESCTQKDVE LLAQQVFVVS TSAPKLPLQI 180
181 EDASRRAPTD EEKASEQENQ LAVVNLDTRL DNRVIDLRTP TSHAIFRIQA GICNQFRNIL 240
241 DVRGFVEIMA PKIISAPSEG GANVFEVSYF KGSAYLAQSP QLYKQMAIAG DFEKVYTIGP 300
301 VFRAEDSNTH RHMTEFVGLD LEMAFNFHYH EVMETIAEVL TQMFKGLQQN YQDEIAAVGN 360
361 QYPAEPFQFC EPPLILKYPD AITLLRENGI EIGDEDDLST PVEKFLGKLV KEKYSTDFYV 420
421 LDKFPLSVRP FYTMPDAHDE RYSNSYDMFM RGEEILSGAQ RIHDADMLVE RAKHHQVDLA 480
481 KIQSYIDSFK YGCPPHAGGG IGLERVTMLF LGLHNIRLAS LFPRDPKRLT P |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
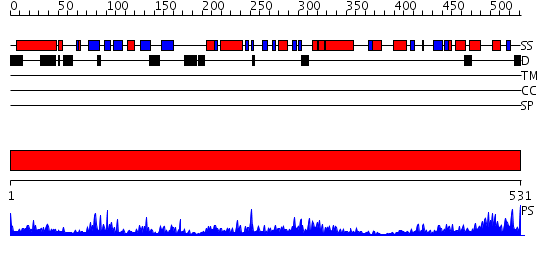
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..531] | 115.0 | Aspartyl-tRNA synthetase (AspRS) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)