













| General Information: |
|
| Name(s) found: |
eme1 /
SPAPB1E7.06c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 738 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC Holliday junction resolvase complex [TAS |
| Biological Process: |
DNA repair
[TAS meiotic joint molecule formation [TAS reciprocal meiotic recombination [IDA replication fork processing [IMP intra-S DNA damage checkpoint [IDA |
| Molecular Function: |
magnesium ion binding
[IEA]
DNA binding [IEA] crossover junction endodeoxyribonuclease activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTLQSTDSAI VIDSEADVDI SSSQASPSKV CRDNIALSEH NVITVLDTPQ RSTQCDSLLK 60
61 SFSTPLVSGS EDVLPSPRDA LNITNKKSVT DNLLLSLTSS NQSTNTNLNP SSRVEIINLN 120
121 SSPPNSLSSQ PKHQEFHLFH TPTIPRTTQL SSKTSSPIVI PDDNEQVASP LSKKAASLTS 180
181 SPLKDFQSSP PLSTVLQKSH SLHDILLDTN DDDHFPFTQS PLTKTKSFND ALTSSSSILK 240
241 PCMPSIASPT SNRLSHAPST PNLFPNQDSS NTIDLINNRS KTSVENQERT FNLTSDVHLD 300
301 SPTSPSKHSS IEPNTSQEDS FQELPSLNKL SIQSRAFKKM RLPKISRTTD TPPASTSNSN 360
361 KKNLDKLKKM RKLCSRSLEP YELDSNTQRK RKRYEDSLKK SKTLDKVDSL NRKMAKELDR 420
421 KNSKELQKIN KVKRTKEECL SEIILLSPDD WASSWYSTVR SQLDSYNCQF VVNSNQPKDS 480
481 IMWKRKVNNV FNSSTNRFEL SIEHEQIEPF ALLRLKCRDF IKYIEEDQAD TFFHEMSEKF 540
541 KGCKLILLLE GIPNYFKSLK AELNRQYAAA VNSGTRPLLF GSLSKYQNFT KEKLESEIVR 600
601 FSFEHSILIN TSNDEKETAQ WIVSFTGDIA LSRYKHFSKF SARASTTEIG HVKSADRIEN 660
661 SLNFMLRQIL RVTPNIANAI CDQFDSIPSL IHHLKTHGEE SLTNVVIQSS ISERNLGPVL 720
721 SRRIYNTFLC KEASSDAP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
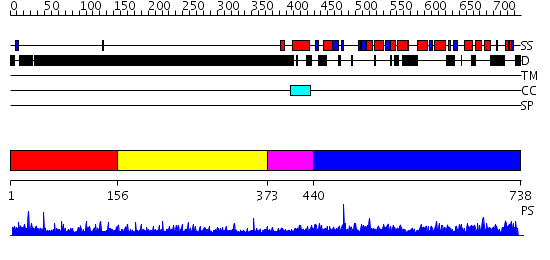
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..155] | 2.225996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [156..372] | 8.244995 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [373..439] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [440..738] | 2.22 | XPF from Aeropyrum pernix, complex with DNA |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)