













| General Information: |
|
| Name(s) found: |
elg1 /
SPBC947.11c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 920 amino acids |
Gene Ontology: |
|
| Cellular Component: |
Elg1 RFC-like complex
[IDA nucleus [IDA chromosome, telomeric region [IEA] nuclear chromatin [IC nuclear replication fork [IC] cytosol [IDA DNA replication factor C complex [ISS |
| Biological Process: |
DNA repair
[IMP DNA-dependent DNA replication [ISS mitotic sister chromatid cohesion [ISS] DNA recombination [ISS telomere maintenance [ISS |
| Molecular Function: |
ATP binding
[IEA]
DNA clamp loader activity [IC |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQIVGYLSAD SQSNPDLKSE NEAKEEKPIG RRHTMSPVPA TSENKYFGKS PLSGSRKPRR 60
61 SRSLHKERSY MRKFFDMDME ESKDFENDQS LLVTLKVSTS LGQKIENILH PKLSNDTNST 120
121 AFPPAKSSGE ASDTNILVEN INSQETVNSS PLVSELHYSN LADSPSNLRN TVTSMHPFFM 180
181 SKSVKKNSEI KFVSEERGGT KPERLLDPLW PTPDSQSMLE YAGSIEPSVF WFPKKHLEEA 240
241 ILEETSHLSF KEVLSSTTAN MITPLAEKNK TEVLQVTPSK LHTFALESLC FSPAPFIQKV 300
301 LSRLLPSDPN VEMPMIPQIL EKGLWVSKYA PSKTQDCCAF SQCLSKIADW LRSCRLTKPE 360
361 SSSVPPSSSI SRSSTIHSCT SSKRNEDSLS ESDFEPDIIE EEDDSDEFNP SVSRKKAKLT 420
421 SSQFSNWMLV TGVTGIGKTS CLYAICRELN FEVVEIHPGM RRSGKELLER IGELTQSHIV 480
481 DKSRLNNTPD ILILLEEVDI LFQDDRGFWQ AVSTLIEKSK RPVVMTCNET DFLPSAFLQE 540
541 DHIVQFQSIS SALLTDYISS VLYADRCIIS RNVVESISYR YGSDLRGILM QLNFWSLVNF 600
601 PSLPSKDKQD DSHEPFIEAT ISAFDEGVGV YNPRIQTSED LLQTYSEEQM GDIGLLFMPN 660
661 LVNWRKVCVP KSEMEEKEAI MEKLIYSHQY ADSLSYVDYR FSSQPTIYET YELMNDSASF 720
721 EDMSLECRDN CANAFQDNLV GFPTISNPFH ANAPPEPHEL KLQYHSFCFI NNLFSKSSLK 780
781 AISSNDSIVP KALNNRELQL SALASTIGYK LDPDDVYNIL SFLSFANSQV TSYTPPNSID 840
841 RPNDILILEV APFVRCMRRY DRIRLNSYKL LLSSKGRSAS HISRRGAASI LRSAGYNYGR 900
901 LQYLEGSDRI LSTWFSTTLD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
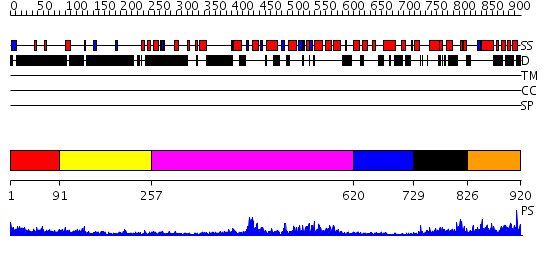
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..90] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [91..256] | 63.69897 | VCP/p97 | |
| 3 | View Details | [257..619] | 6.522879 | No description for 2c9oA was found. | |
| 4 | View Details | [620..728] | 29.045757 | ADP Bound E. coli Clamp Loader Complex | |
| 5 | View Details | [729..825] | 59.0 | EDTA treated | |
| 6 | View Details | [826..920] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)