













| General Information: |
|
| Name(s) found: |
spc7 /
SPCC1020.02
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1364 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cell division site [IDA NMS complex [IDA spindle pole body [IDA condensed chromosome, centromeric region [IDA cytosol [IDA inner kinetochore of condensed nuclear chromosome [IDA |
| Biological Process: |
attachment of spindle microtubules to kinetochore
[IGI chromosome segregation [IMP response to pheromone [IMP cellular protein localization [IGI cell cycle [RCA |
| Molecular Function: |
protein binding
[IPI microtubule plus-end binding [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPTSPRRNSI ATTDNVIGRN KSRKRPHSLG GPGALQELKE HTNPAKGILK SYSSFSVDPA 60
61 FTGDEDFNDI HTQINNTVIG ISSNVMAASK RLQMEDLQQL SRETSRKSLN RRVSFASHAR 120
121 VRWYPKDHQS DSEKSTSNHS TPERTFASDA KNHSPKGPTT TSFSRNETQS SPHSHSASII 180
181 SDGSDMDIAS PIRSTESDMV SEALNAGHPP PSLYPENDDL SIQNPTKALP EAEKALDVHD 240
241 ATREQVNDRE ETNMDLTIQF QEADSFLSHS ESIKGLSSSE QGTVYSLKAS HDPSNQTQLS 300
301 SPNKSSSPTS IEISDFSKNN ENHDQSENKE EEEDMMLTRP IEIPQHFSPI ARPLTSQEAI 360
361 VDMDITSNNI NLSPVSHFSN GLDLQNLEEA PMNLTRPINA NPHLTNHSPN DLTNGEEEMD 420
421 TTSAFNIENS HLTLLSPIRP SSRSMEEQIM DLTQPISSTN APTHLNEDDL NQFTSNISSS 480
481 SKPRKDNNKT ANSSKPIPDS EDFMDITRPF NILSPSKEAL SEEQPMELTS TVFPCENSTS 540
541 HLEVEEAAMD ETVAFQIRGN NVELPSADKE NAEREEIPSY SDKSENFNTT SFTNHERSPN 600
601 GNNNLKFSKD PNSSSPSRHV VATPTDKLGT RKRRLRYSTS SFDQSTLRRN RLATIRNARK 660
661 SISTLNDREL LPVNFFEKKV NSGLYKSVER SENYRLGATP LTAEKPFTTE KPLSSLPEEV 720
721 SRQPTDDKGE QVSNADVDSG LSKTERLTIQ QTNEIKHVPT NTTSSVKLPQ QPSNEDEKER 780
781 ITTADYADST SLERLESQEP NRNELVQVGS SNAGNTTSVG MNEHEKSPVK LSKGVSNVDT 840
841 SLGASTINTN ILNQDSGPNE EIPVGNEPEF DTMPTLPNVE PISLSDFLKM TGIEFLDNLT 900
901 IAKRRETLLP NAEENKKCSI QELLESFYIQ FPLLELYKFS CQQLQDYIAE GKDFVTKIEE 960
961 ETLKENPLLF YEYRKASSDM RVLMDSQFLM MKTFARLQAK GDWYEWREGL MQGIKHELNL1020
1021 NLTGMQRSLT HLMDVANVIH PYAQEIQERY NGSITTVQTL KKQKEFANQY DSTLLAQAQE1080
1081 KLEKLKVEVE RRRRLLSEKE ERRKELAIKI EQVTNSCSDL ELRTNAEQDF YAKNQDFEFD1140
1141 EIKRYEEQLL NLKNELGWTI VSLTAGGIKL ATNNTALSPY SAEVTVEILR QNFQVNVDIA1200
1201 CKFPNESNAC SSNVLEHVAS SFSKWHSKVF SRNLRLLKKY LNDVSICWEQ IVYLVQDFQR1260
1261 LWYHWPFLSV ENDDKSIIIN VELYLRSVSS KVKVVFGLPI DTIYQTTEVG KFYASTSVAV1320
1321 KQMYAESEGD SYVSEVLNTL SEVVHCTSTY ALSSACLTVW NKYS |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Liu X, et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
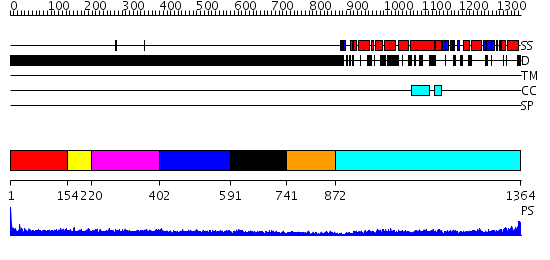
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..153] | 1.201994 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [154..219] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [220..401] | 3.218996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [402..590] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [591..740] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [741..871] | 15.09691 | No description for 2dfsA was found. | |
| 7 | View Details | [872..1364] | 7.39794 | Crystal Structure Analysis of ClpB |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)