













| General Information: |
|
| Name(s) found: |
snd1 /
SPCC645.08c
[Sanger Pombe]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 878 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA RNA-induced silencing complex [IEA] |
| Biological Process: |
RNA interference
[IEA]
sphingolipid biosynthetic process [NAS] |
| Molecular Function: |
hydrolase activity, acting on ester bonds
[IEA]
RNA binding [RCA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQYVSSMIK YAQSGDSFNI LIKDNAKKIT EKQFSLAYVE CPRFRREGDE PFAFEAQEFS 60
61 RRLVVGRPAS VSTLYVIPTS KREYGRIRTS EFDLAESLLR EGLAKLRPEA TRNEGTSENS 120
121 YFVSLEEAQD HAQQYKLGIW GPSDDVVVTE KANPANPAKF LKAHKGKKLN GIVETIRNGD 180
181 QVRVRLFLSP KQHQLVTISL AGVRCPRSTF TATSPEQTSS EQEPCGDEAK QFVVTRLLQR 240
241 NVVIELLDLA PNGVSFLGNV LHPAGNIATF LLSSGLGRVA DNHISALGPE TMQSLRTIER 300
301 KAKISRLGIW KNISVSIPDI NSLSLKDYSA VVSRVISTDT LEVRKDNGVE CRIQLSSIRH 360
361 PRPSNEKEAP YQLEAREFLR KKIIGKRVQV SLDFIRPGQN DLPAINNCTV KLSDGTNVAL 420
421 MVVKSGYATV IRYRMDSVDR SPIYDFLIEA EKAAQEGRKG MWSGKKPAYE NIVNASESSL 480
481 RSRQYLSSLQ RTRKLSVIIE NVISGSRFRC FCPKENCYFM FACAGIRTPR TARNDQEKGE 540
541 PFAEESLSLA KSLLQHDAQV EILSVDNNGC FLGDIYVNHD TNFALKLLSQ GLAWCQGYAS 600
601 QSNVQYSQYH DTEAAAKEQK VGMWHDYVPP EKKAASTEKE SENTVKEPIY LDIVLSDIAE 660
661 DGKFSFQIIG TGIQQLETLM SDLGSLKKSF KPSEKINVGM NVAAISALDN AMYRGRVLRC 720
721 DRENQAADVL LYDYGSVEQI PFKNISSLPD TYTKLKPQAQ LARLSYVQLP PPSSDYYEDA 780
781 RLVFRELAMN KGLVAKVDGH EGNVYSVTLY NPSDGSDFSD CINAQLVALG MASVIPKKKT 840
841 SHFEKDTASL NILEEHQQEA RLNHIGFWVY GDPLEYED |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
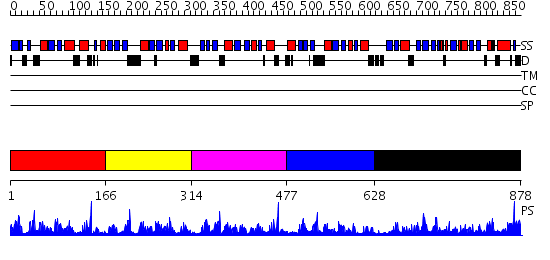
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..165] | 27.522879 | No description for 2f0uA was found. | |
| 2 | View Details | [166..313] | 19.69897 | Solution structure of 1-110 fragment of Staphylococcal Nuclease with G88W mutation | |
| 3 | View Details | [314..476] | 15.853872 | No description for PF00565.8 was found. No confident structure predictions are available. | |
| 4 | View Details | [477..627] | 4.63 | ACCOMMODATION OF INSERTION MUTATIONS ON THE SURFACE AND IN THE INTERIOR OF STAPHYLOCOCCAL NUCLEASE | |
| 5 | View Details | [628..878] | 54.522879 | No description for 2hqeA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)