













| General Information: |
|
| Name(s) found: |
usp104 /
SPAC4D7.13
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 695 amino acids |
Gene Ontology: |
|
| Cellular Component: |
commitment complex
[ISS nucleus [IDA U1 snRNP [IDA |
| Biological Process: |
nuclear mRNA 5'-splice site recognition
[IC]
nuclear mRNA splicing, via spliceosome [IPI |
| Molecular Function: |
RNA binding
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSAPPWQTSE YDETEGFTSN QEGPSAAPSK TVASDWHEVK TEDSRVYYYN SVTRKSVWEK 60
61 PEELMNDFEK KLSKLAWKEY ATADGKKYWY NVNTRESVWD IPDEYKAALV DEPEQQKKAL 120
121 SSKIKSNDNK PAVQSIQRHG PDVAAPSSQP AKDQSQQISQ GSHKRTINFV QQKDKRQKRS 180
181 NDYQHENYDT YEAAERAFFK FLDSHNVNPS WTWEQTVREL CDAKGYYVMK DPWHRKCAFD 240
241 AYILNYLTDQ SDAEKNRVTK IRKEFIEMLK SSDKIHSYTL WRTVKNEFSS HPAFNATSSE 300
301 TEQQQLFFEY KQKLLEDEKQ LEKDRRKEAL DDFCSLLRNM NFEPYTRWSV AQAKFDQDPR 360
361 YTRNSNMKYL SKLDALVAFE DHVKHLEREY ILDKQKQKKE KHRIERKNRD AFRALLQDLR 420
421 VQKKITLRTK WKELYPIIKD DPRYLNLLGQ SGSTPLDLFW DTIVDLENMY REKRNLVLDC 480
481 LEVLQISVDD TSNIPEIIAR LSEKLKDREE SEAVTEDLIE EVVNRLRDKA IHKKAEEKRA 540
541 DERRIRRKID NLRSAIKYLK PPISADASYD EIRPLISILP EFAALHSEEH RMAAFDKYIR 600
601 RLREKRELEK QYQNRRGYYD VGKDESYLAN SARPHSGYED GRLEYSADLA SKSNRNEINT 660
661 MQDVQENSIS HVTATQPAVK NIVDDAESSE EGEIR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
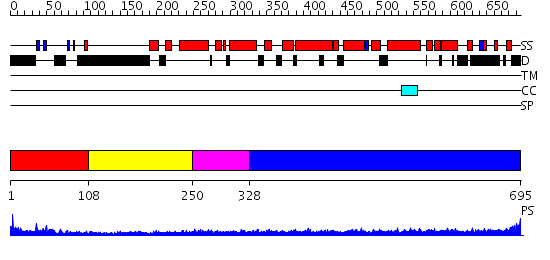
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..107] | 4.221849 | Splicing factor prp40 | |
| 2 | View Details | [108..249] | 2.77 | No description for 2doeA was found. | |
| 3 | View Details | [250..327] | 2.21 | Solution structure of the FF domain of human Formin-binding protein 3 | |
| 4 | View Details | [328..695] | 11.69897 | Heavy meromyosin subfragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)