













| General Information: |
|
| Name(s) found: |
SPBC3B9.03
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 547 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA signal recognition particle receptor complex [ISS |
| Biological Process: |
SRP-dependent cotranslational protein targeting to membrane
[IC |
| Molecular Function: |
GTP binding
[ISS GTPase activity [ISS signal recognition particle binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIDLFAIVNK GGIVLWKKTN SLVNLKCLQV LFHEAFLSEQ RTVNNTVTFD RYTMQYQEAT 60
61 QYSIVFVVVF QDLKCMAYSQ SLLNSAHNIF LNLFKEKLED RQVPNEAEVE KLFAPIFNRK 120
121 SAQLENETDT KSLPVEANND NSARKKNEYE MKKKGAQSKQ TNAPKKGKKQ LRKWDDQITE 180
181 EEQAALNYSS QASSASQTVD NSQLSSIVGN NNKFQKTGSG DVIISDLEMD PNQTISNKSA 240
241 SSAFSLFSNL IGGKYLKEED LSPILKQMQE HLTKKNVANS IALELCESVK ASLINKKVGS 300
301 FDTVKNTVNK AFRDRLTQIL TPSTSLDLLH SIRSVRKNEN RPYTISLIGV NGVGKSTTLA 360
361 KIAYWLLSNN FRILVAACDT FRSGAIEQLG VHVKNLQSLK GSSIELFAQG YGKDSSFVVK 420
421 NAVEYAKQNS FDVILIDTAG RRHNDQRLMG SLEKFTKATK LDKIFQVAEA LVGTDSLAQA 480
481 KHFQASLYHR PLDGFIISKV DTVGQLVGVM VGMVYAVRVP IIFVGIGQTY SDLRTLSVDW 540
541 VVDQLMK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
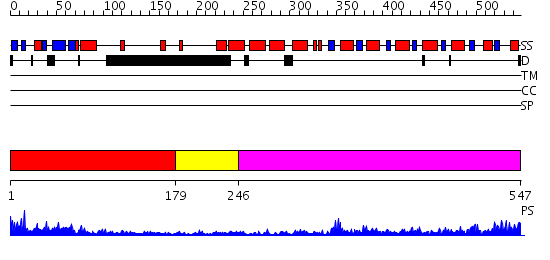
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..178] | 24.154902 | The Structure of the Mammalian SRP Receptor | |
| 2 | View Details | [179..245] | 11.522879 | N-terminal, ClpS-binding domain of ClpA, an Hsp100 chaperone; ClpA, an Hsp100 chaperone, AAA+ modules | |
| 3 | View Details | [246..547] | 54.0 | No description for 2j289 was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)