













| General Information: |
|
| Name(s) found: |
pub2 /
SPAC1805.15c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 671 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA cell cortex of cell tip [IDA cytosol [IDA extrinsic to membrane [IEA] |
| Biological Process: |
protein ubiquitination
[RCA |
| Molecular Function: |
ubiquitin-protein ligase activity
[IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MENIRFEVQL TILHVEGLWK NGLLRSLKPY LLISVDDDQF IKTNVASGTL RLSWGFTQKL 60
61 TVSPQSIILL QLFDEKQKNE TSDGFVGLGA AVVNSFLPFN NPKDDYKTRI TLRSPSGSYR 120
121 GSVVCLFKRS KFLPEELPAD KSQICTDIID DASGCAWETR IDEFGHVYYL KSPQLSVISA 180
181 ISHEKLENLT PKQLKEVFSQ FLFNNQSKSS LKINLEYKVI KHLLEHYPLA LSVRQQVAVE 240
241 KGPLPAGWEM RLSEDYHVYF VDHSTKTTTW SDPRDNVVAS DSVSENTDSI QQINDEYQRK 300
301 IAYMYDRPEM AVNDAQLQLK VSRATTFEDA YDIISKLSVS DMKKKLLIRF RNEDGLDYGG 360
361 VSREFFYILS HAIFNPGYSL FEYATDDNYG LQISPLSSVN PDFRSYFRFV GRVMGLAIYH 420
421 RRYLDVQFVL PFYKRILQKP LCLEDVKDVD EVYYESLKWI KNNDVDESLC LNFSVEENRF 480
481 GESVTVDLIP NGRNIAVNNQ NKMNYLKALT EHKLVTSTEE QFNALKGGLN ELIPDSVLQI 540
541 FNENELDTLL NGKRDIDVQD WKRFTDYRSY TETDDIVIWF WELLSEWSPE KKAKLLQFAT 600
601 GTSRLPLSGF KDMHGSDGPR KFTIEKVGHI SQLPKAHTCF NRLDIPPYNS KEELEQKLTI 660
661 AIQETAGFGT E |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
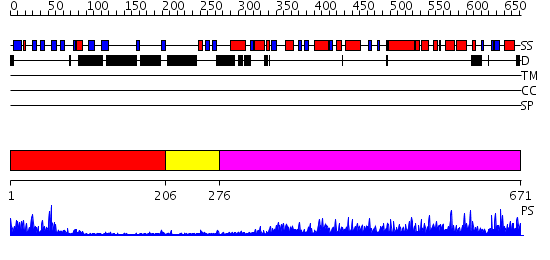
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..205] | 22.69897 | Synaptogamin I | |
| 2 | View Details | [206..275] | 2.87 | Splicing factor prp40 | |
| 3 | View Details | [276..671] | 135.0 | Conformational Flexibility Underlies Ubiquitin Ligation Mediated by the WWP1 HECT domain E3 Ligase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)