













| General Information: |
|
| Name(s) found: |
tsc2 /
SPAC630.13c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1339 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA cytosol [IDA TSC1-TSC2 complex [IPI |
| Biological Process: |
regulation of small GTPase mediated signal transduction
[IEA]
regulation of amino acid import [IMP regulation of GTPase activity [IC regulation of L-arginine import [IGI TOR signaling pathway [TAS regulation of intracellular protein transport [IMP cellular protein localization [IGI positive regulation of GTPase activity [IEA] cellular response to nitrogen starvation [IMP |
| Molecular Function: |
protein binding
[IPI GTPase activator activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNNKSLLDLS SEPAIKDTDI LSYFDANLPF KKRHQIVRSI YHGLKYYIYS STSIIQVWQN 60
61 IQDFISTKND NAAFRELVYN LMMRYVSTQK HLSIIERHTF FQTIEKDPFQ DIAELQLRLK 120
121 SLSVLTDEGH KISGIENRVG PLLSAWFNQY LQWQSQATEL QGKDADSKLV HLLFFKSLFK 180
181 FSTNLVKFQW FLVPEPQMLQ LVNSVVQICN HARLEDVVTE VLMFFDSMIR YSVIPKASLY 240
241 DTVLILCSTY ISTYSYSKLA QSVIFNLISS PVSNLAFENV FNILQYNRSN VNAVRGAVRL 300
301 MRFLMLQEVK NDAIASITLS SSIEFTEFPL GFNENVDFEI LGTVYLFLRT PSILNRLNFL 360
361 DWHRILNILM YCSQYLPLKA STSKEAFSKT AAFANIYDRV LDFLDFEALI PLLQQFQVKL 420
421 VFFLKDVLPV LKPKIRKKLL RLFETYNLIF PCNQYWVFNL EFLLGIYQCK TFDLEDRALL 480
481 FKLVEDACSV ADENSAPILC SKFLFPVIES FSKESDDCVV SPVYNMLFFL SVNFQNPGLK 540
541 DCIDHIFQQL ISDTSSVTVR RLATSTLIRL FYYYYDLRDA VPIQETLAKM LEILETPSFP 600
601 FVSRMLCLQF FLRFRANGTS IYICENIDLN EPFKVLNVDS ELIPAVYPIS DSFVNSATVE 660
661 KHIWERKEND LIKISNHSTE YGKDFVTFPT SSLLRFYRKS MATESNWTIL MFMITHLADQ 720
721 ISNRSMFIGA LEEIYNLLDF MCDIVFERVS ISAEIPSNIR KANIMIPILQ NVQMLFVYHD 780
781 QFSRAQEDEL VSVFFAGLQK WNEACHVSIH SLMLCCYELP VSIRKQLPAI LVTLSRLITK 840
841 PDLSVHILEF LCSLARLPDL IANFTDADYR QIFAIALKYI QHRDFTKESK DSNDTESILK 900
901 NSYSSYVLAL AYSVLQIWFL SLRLTERKKF VPWILRGLKL ASEGKPLEDL CLVQYDMMQQ 960
961 FCYSNSDINN QTSTFVDSDV ESETWIRGNS LFTINVSVNS GFLEAVIRRP TGTTQYTFRN1020
1021 EASLQKFLWE ENLTSSKALT RGLLCTPSSF VSHFLDPHGI SLYNQPLLLP SNDDSVRRAI1080
1081 SVFDRIPVIE SLKAGLVYVG YQQRREADIL ANTNPSEDFL TFLNGLGTLF ELKTDQKVFA1140
1141 GGLDRENDID GAFAYCWKDK VTQMVFHCTT MMPTNIEHDP GCTLKKRHIG NDFVTIIFNE1200
1201 SGLEYDFDTI PSQFNFVNIV ITPESESIRR TGRQIKFYKV KALTKYDIDF SLFRRYKIVS1260
1261 SDALPAIVRD VTLNAAVFSH IYHRSAGDYV HIWAERLRQL KRLREKFQAS VLPEDYNLDE1320
1321 QTKTKLQNGT NFSDFTSYL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
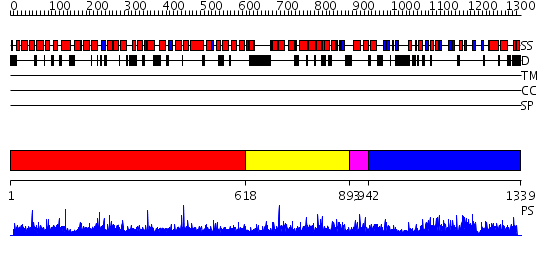
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..617] | 0.966 | Adaptin beta subunit N-terminal fragment | |
| 2 | View Details | [618..892] | 0.968 | No description for 2ie3A was found. | |
| 3 | View Details | [893..941] | 1.039992 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [942..1339] | 91.154902 | Crystal Structure of the Rap1GAP catalytic domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)