













| General Information: |
|
| Name(s) found: |
bpb1 /
SPCC962.06c
[Sanger Pombe]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 587 amino acids |
Gene Ontology: |
|
| Cellular Component: |
commitment complex
[IDA nucleus [IDA cytosol [IDA |
| Biological Process: |
generation of catalytic spliceosome for first transesterification step
[IDA |
| Molecular Function: |
zinc ion binding
[IEA]
RNA binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLNSRSVGST GSNNTPLGRR RFDEKPDSLP PLPDANGMSN GYRDSYKSNS RMDHRPDGYH 60
61 DGRGRRAYRK HYWGHPTPIE EMLPSQMELE TAVKSCMTME QLELYSLNVR LEEITQKLRT 120
121 GDVVPHHRER SPSPPPQYDN HGRRLNTREI RYKKKLEDER HRIIERAMKM VPGFRAPSDY 180
181 RRPAKTQEKV YVPVKDYPEI NFIGLLIGPR GHTLKDMEAK SGAKIAIRGK GSVKEGKGRS 240
241 DPSVRGNMEE DLHCLVTADS EDKINHAIKL IDNVIQTAAS VPEGQNDLKR NQLRQLATLN 300
301 GTLRDDENQV CQNCGNVGHR RFDCPERINH TMNIVCRHCG SIGHIARDCP VRDQQPPMAD 360
361 STADREYQSL MQELGGGSAI SNGNGEPQKS IEFSESGAAS PQAGHPPPWA AASTSVSSST 420
421 SSPAPWAKPA SSAAPSNPAP WQQPAAPQSA PALSMNPSSL PPWQQPTQQS AVQPSNLVPS 480
481 QNAPFIPGTS APLPPTTFAP PGVPLPPIPG APGMPNLNMS QPPMVPPGMA LPPGMPAPFP 540
541 GYPAVPAMPG IPGATAPPGA PGSYNTSESS NLNAPPGVSM PNGYSNR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
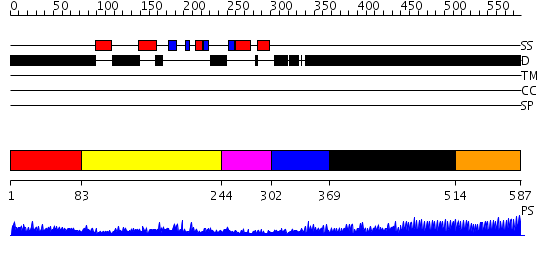
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..82] | 3.064993 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [83..243] | 50.154902 | HIV capsid protein, dimerisation domain; HIV-1 capsid protein | |
| 3 | View Details | [244..301] | 17.39794 | RNA splicing factor 1 | |
| 4 | View Details | [302..368] | 12.0 | HIV nucleocapsid | |
| 5 | View Details | [369..513] | 1.13 | No description for 2j63A was found. | |
| 6 | View Details | [514..587] | 1.8 | Crystal structure of the C-terminal domain of BclA, the major antigen of the exosporium of the Bacillus anthracis spore. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)